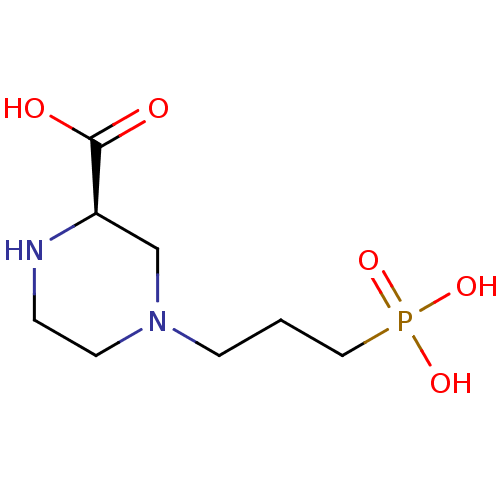
BDBM50050704 (R)-4-(3-Phosphono-propyl)-piperazine-2-carboxylic acid::(R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid::CHEMBL47277
SMILES C1CN(C[C@@H](N1)C(=O)O)CCCP(=O)(O)O
InChI Key InChIKey=CUVGUPIVTLGRGI-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 50050704
Found 3 hits for monomerid = 50050704
Affinity DataKi: 41nMAssay Description:Inhibitory constant against (S)-glutamate and glycine evoked currents mediated by N-methyl-D-aspartate glutamate receptor NR1a/NR2A expressed in Xeno...More data for this Ligand-Target Pair
Affinity DataKi: 270nMAssay Description:Inhibitory constant against (S)-glutamate and glycine evoked currents mediated by N-methyl-D-aspartate glutamate receptor NR1a/NR2B expressed in Xeno...More data for this Ligand-Target Pair
Affinity DataKi: 630nMAssay Description:Inhibitory constant against (S)-glutamate and glycine evoked currents mediated by N-methyl-D-aspartate glutamate receptor NR1a/NR2C expressed in Xeno...More data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)