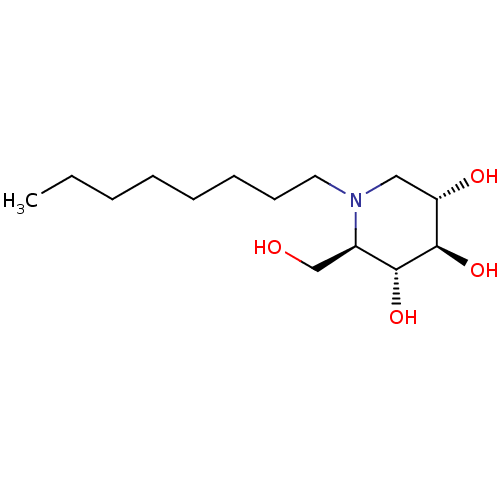
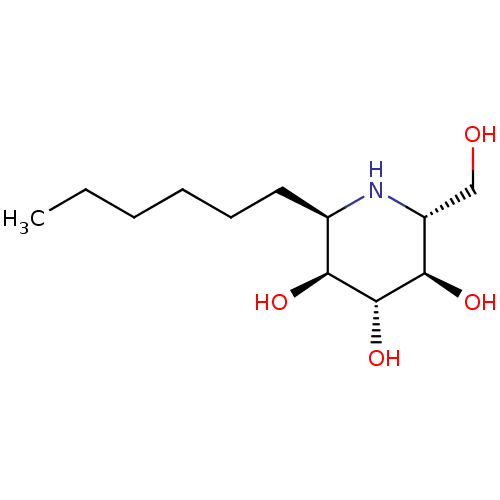
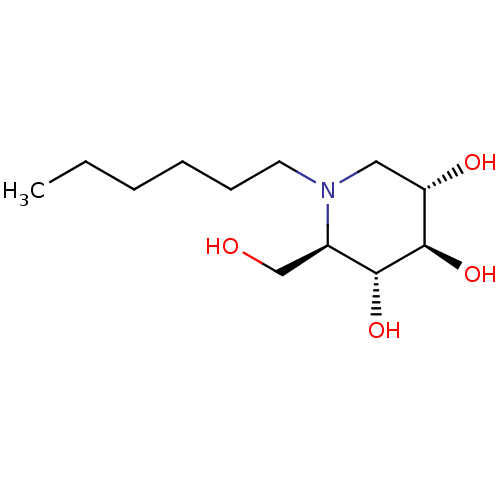
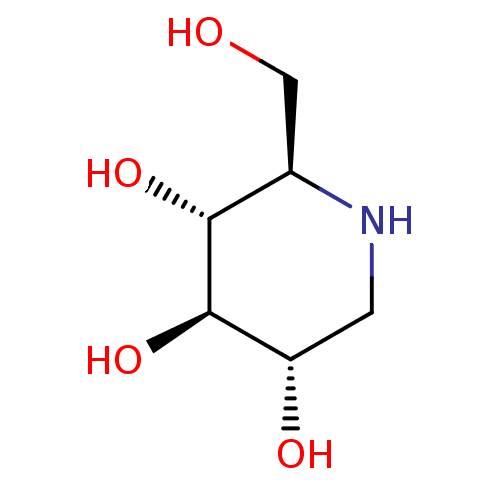
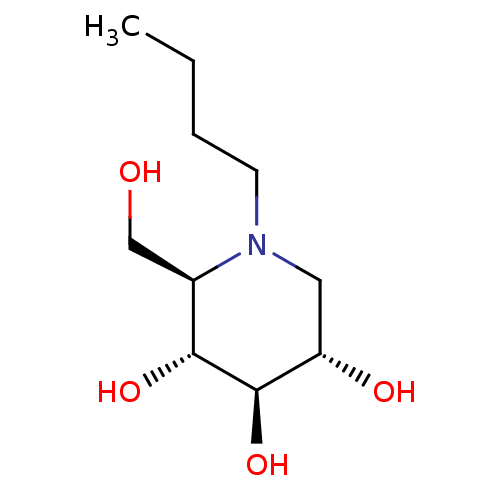
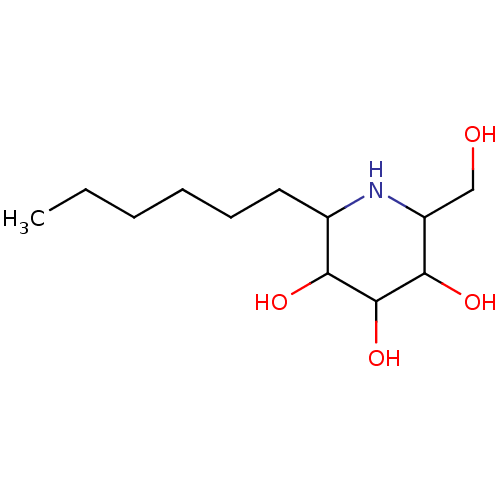
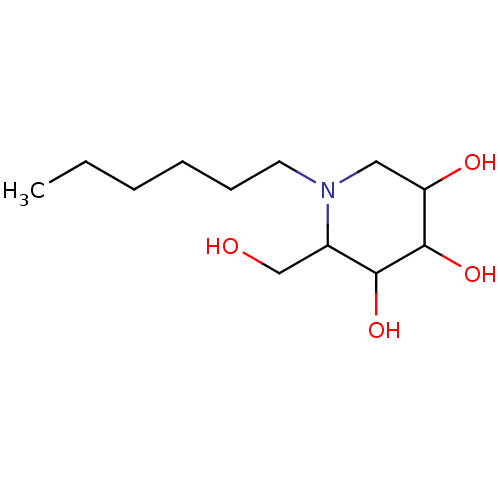
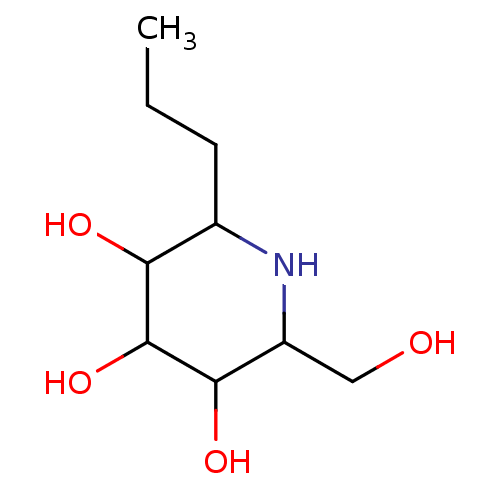
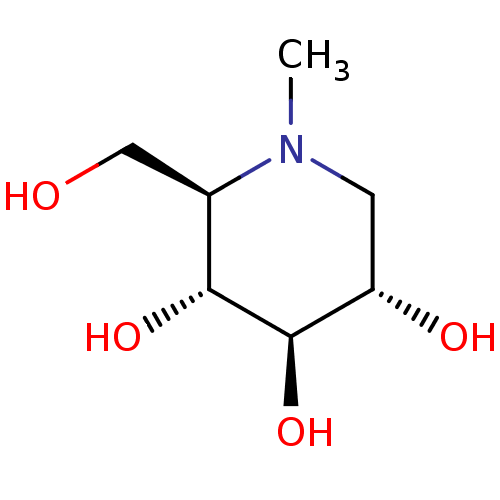
Affinity DataKi: 200nM ΔG°: -39.8kJ/mole IC50: 270nMpH: 5.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
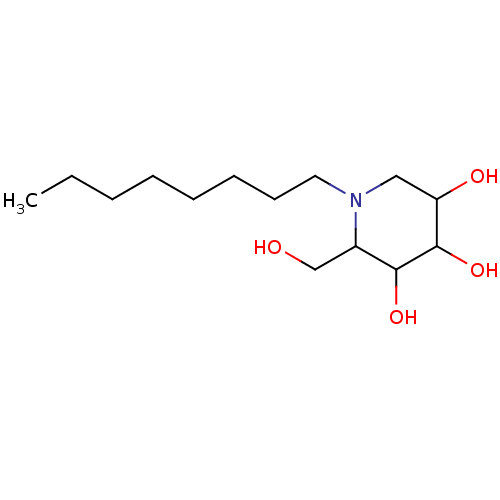
Affinity DataKi: 280nM ΔG°: -38.9kJ/mole IC50: 500nMpH: 5.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
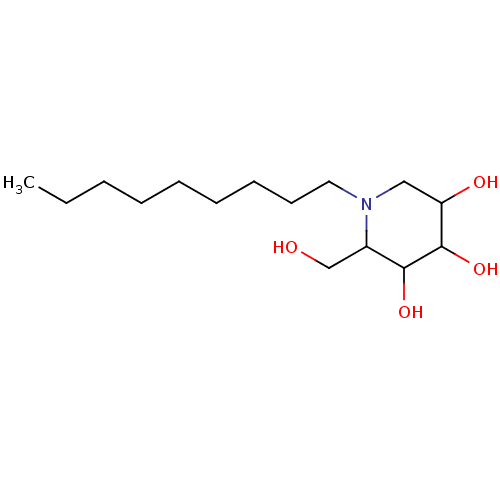
Affinity DataKi: 300nM ΔG°: -38.7kJ/mole IC50: 660nMpH: 5.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
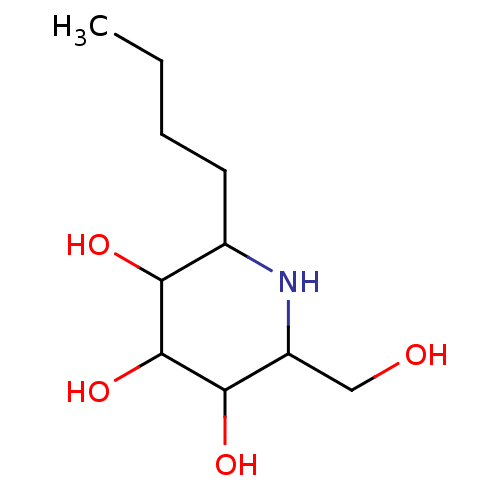
Affinity DataKi: 420nM ΔG°: -37.9kJ/mole IC50: 820nMpH: 5.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
Affinity DataKi: 2.30E+3nM ΔG°: -33.5kJ/mole IC50: 4.20E+3nMpH: 5.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
Affinity DataKi: 5.50E+3nM ΔG°: -31.2kJ/mole IC50: 1.30E+4nMpH: 5.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
Affinity DataKi: 7.90E+4nM ΔG°: -24.4kJ/mole IC50: 2.40E+5nMpH: 5.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
Affinity DataKi: 1.10E+5nM ΔG°: -23.5kJ/mole IC50: 1.00E+5nMpH: 5.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
Affinity DataKi: 1.16E+5nM ΔG°: -23.4kJ/mole IC50: 2.70E+5nMpH: 5.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
Affinity DataIC50: 3.5nMAssay Description:Inhibition of rat intestinal isomaltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 50nMAssay Description:Inhibitory activity against alpha-Glucosidase from riceMore data for this Ligand-Target Pair
Affinity DataIC50: 80nMAssay Description:Inhibitory activity against alpha-Glucosidase from riceMore data for this Ligand-Target Pair
Affinity DataIC50: 80nMAssay Description:Inhibitory activity against alpha-Glucosidase from riceMore data for this Ligand-Target Pair
Affinity DataIC50: 90nMAssay Description:Inhibition of rat intestinal isomaltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 150nMAssay Description:Inhibition of rat intestinal isomaltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 210nMAssay Description:Inhibitory activity against rat intestinal sucrase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 230nMAssay Description:Inhibition of rat intestinal isomaltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 280nMAssay Description:Inhibition of rat intestinal isomaltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 300nMAssay Description:Inhibitory activity against rat intestinal isomaltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 310nMAssay Description:Inhibition of rat intestinal isomaltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 360nMAssay Description:Inhibitory activity against rat intestinal maltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 420nMAssay Description:Inhibitory activity against alpha-Glucosidase from riceMore data for this Ligand-Target Pair
Affinity DataIC50: 450nMAssay Description:Inhibition of rat intestinal isomaltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 450nMAssay Description:Inhibition of rat intestinal sucrase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 530nMAssay Description:Inhibition of rat intestinal isomaltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 590nMAssay Description:Inhibitory activity against alpha-Glucosidase from riceMore data for this Ligand-Target Pair
Affinity DataIC50: 660nMAssay Description:Inhibition of rat intestinal sucrase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 660nMAssay Description:Inhibition of rat intestinal sucrase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 700nMAssay Description:Inhibitory activity against alpha-Glucosidase from riceMore data for this Ligand-Target Pair
Affinity DataIC50: 800nMAssay Description:Inhibitory activity against alpha-Glucosidase from riceMore data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+3nMpH: 4.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+3nMpH: 4.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+3nMpH: 4.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
Affinity DataIC50: 1.20E+3nMAssay Description:Inhibition of rat intestinal sucrase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 1.30E+3nMAssay Description:Inhibition of rat intestinal maltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 1.40E+3nMAssay Description:Inhibition of rat intestinal isomaltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+3nMpH: 4.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+3nMAssay Description:Inhibition of rat intestinal maltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+3nMAssay Description:Inhibitory activity against alpha-Glucosidase from riceMore data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+3nMAssay Description:Inhibition of rat intestinal sucrase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 1.90E+3nMAssay Description:Inhibition of rat intestinal maltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+3nMAssay Description:Inhibitory activity against alpha-Glucosidase from riceMore data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+3nMAssay Description:Inhibition of rat intestinal sucrase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 2.10E+3nMAssay Description:Inhibition of rat intestinal maltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 2.10E+3nMpH: 4.5 T: 2°CAssay Description:Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 362 nm, emission 4...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+3nMAssay Description:Inhibition of rat intestinal sucrase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 2.70E+3nMAssay Description:Inhibition of rat intestinal isomaltase using disaccharideMore data for this Ligand-Target Pair
Affinity DataIC50: 3.50E+3nMAssay Description:Inhibition of rat intestinal maltase using disaccharideMore data for this Ligand-Target Pair

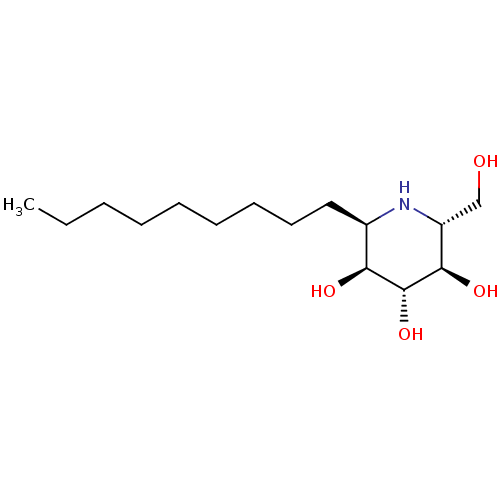
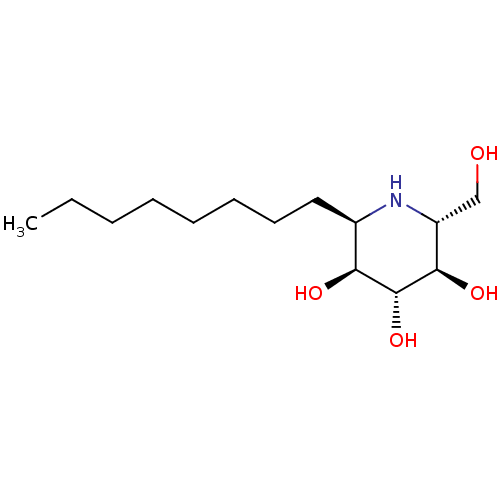
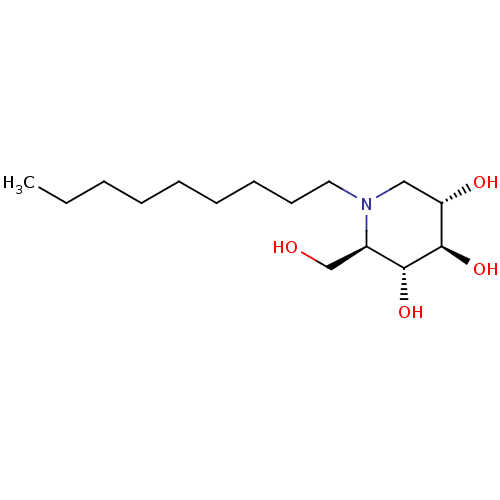
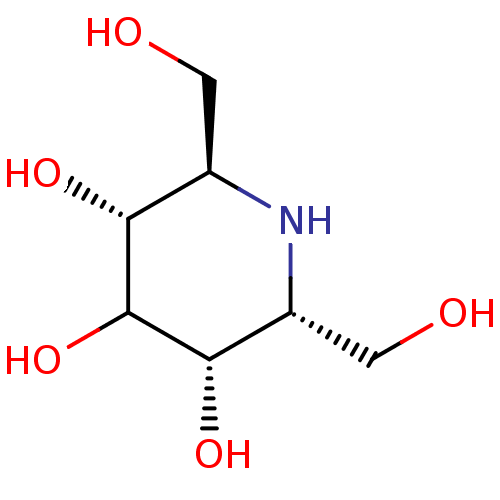
 3D Structure (crystal)
3D Structure (crystal)