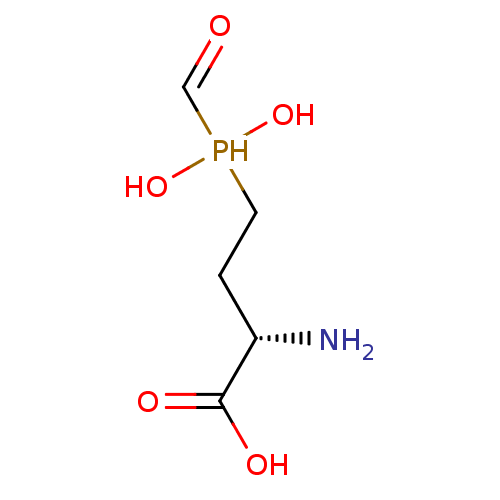
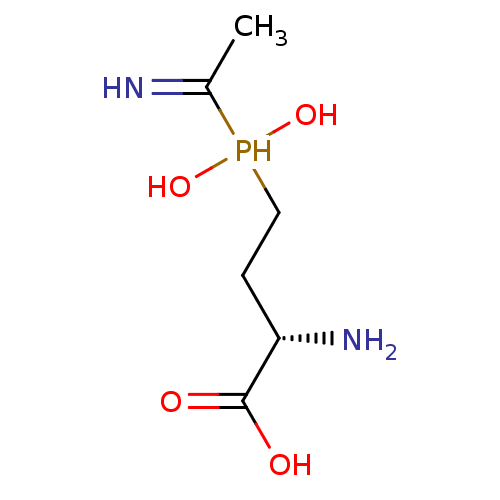
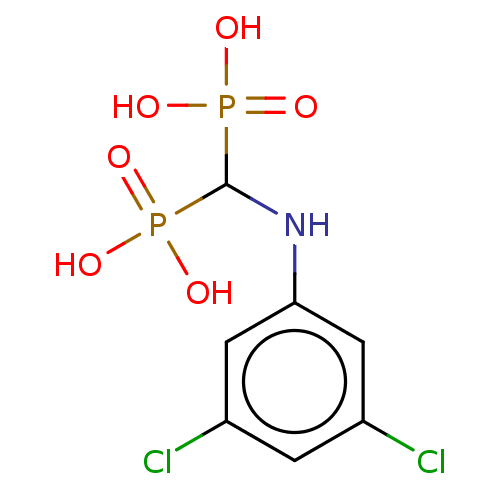
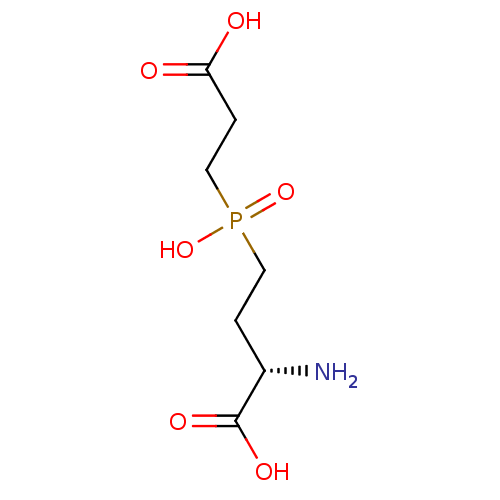
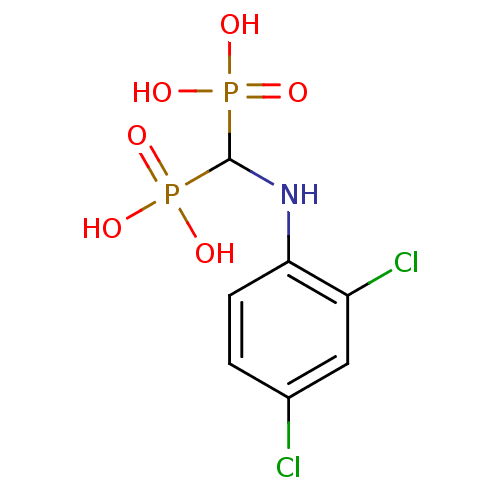
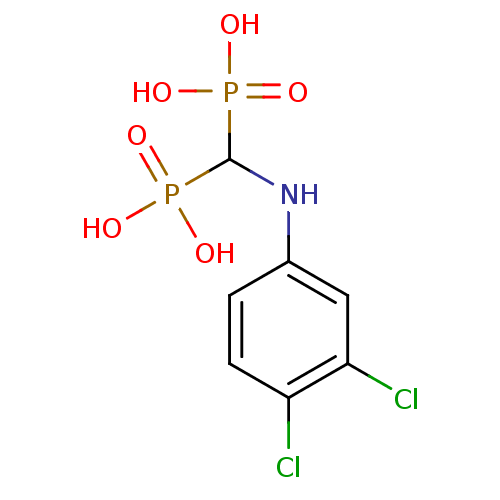
Affinity DataKi: 0.220nMpH: 7.0Assay Description:The enzyme was assayed at pH 7.0 using the pyruvate kinase/lactate dehydroenase coupling system.More data for this Ligand-Target Pair
Affinity DataKi: 0.470nMpH: 7.0Assay Description:The enzyme was assayed at pH 7.0 using the pyruvate kinase/lactate dehydroenase coupling system.More data for this Ligand-Target Pair
Affinity DataKi: 5nMpH: 7.0Assay Description:The enzyme was assayed at pH 7.0 using the pyruvate kinase/lactate dehydroenase coupling system.More data for this Ligand-Target Pair
Affinity DataKi: 45nMpH: 7.0Assay Description:The enzyme was assayed at pH 7.0 using the pyruvate kinase/lactate dehydroenase coupling system.More data for this Ligand-Target Pair
Affinity DataKi: 140nMpH: 7.0Assay Description:The enzyme was assayed at pH 7.0 using the pyruvate kinase/lactate dehydroenase coupling system.More data for this Ligand-Target Pair
Affinity DataKi: 310nMpH: 7.0Assay Description:The enzyme was assayed at pH 7.0 using the pyruvate kinase/lactate dehydroenase coupling system.More data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataKi: 590nMAssay Description:Binding affinity towards glutamine synthetaseMore data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataKi: 600nMAssay Description:Binding affinity towards glutamine synthetaseMore data for this Ligand-Target Pair
Affinity DataKi: 850nMAssay Description:Compound was tested for binding affinity against Glutamine synthetaseMore data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataKi: 2.10E+3nMAssay Description:Binding affinity towards glutamine synthetaseMore data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataKi: 3.40E+3nMAssay Description:Binding affinity towards glutamine synthetaseMore data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataKi: 3.70E+3nMAssay Description:Binding affinity towards glutamine synthetaseMore data for this Ligand-Target Pair
Affinity DataKi: 6.40E+3nMAssay Description:Un-competitive inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using ATP as substrate Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 7.20E+3nMAssay Description:Non-competitive inhibition of Zea mays cv. Jeff( maize) leaf glutamine synthetase using L-glutamate as substrate Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataKi: 1.30E+4nMAssay Description:Binding affinity towards glutamine synthetaseMore data for this Ligand-Target Pair
Affinity DataKi: 2.10E+4nMAssay Description:Inhibition of soybean glutamine synthetase by transferase assayMore data for this Ligand-Target Pair
Affinity DataKi: 4.00E+4nMAssay Description:Inhibition of Salmonella typhimurium glutamine synthetase by transferase assayMore data for this Ligand-Target Pair
Affinity DataKi: 5.30E+4nMAssay Description:Non-competitive inhibition of Zea mays cv. Jeff( maize) leaf glutamine synthetase using L-glutamate as substrate Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 5.50E+4nMAssay Description:Un-competitive inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using ATP as substrate Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 7.50E+4nMAssay Description:Compound was evaluated for binding affinity by the inhibition of Glutamine synthetase (Escherichia coli) activity using biosynthetic assay [mixed Non...More data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataKi: 1.50E+5nMAssay Description:Binding affinity towards glutamine synthetaseMore data for this Ligand-Target Pair
Affinity DataKi: 2.50E+5nMAssay Description:Compound was evaluated for binding affinity by the inhibition of Glutamine synthetase (Pea seed) activity using biosynthetic assay [noncompetitive in...More data for this Ligand-Target Pair
Affinity DataKi: 7.50E+5nMAssay Description:Compound was evaluated for binding affinity by the inhibition of Glutamine synthetase (Escherichia coli) activity using biosynthetic assay [mixed Non...More data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 1.00E+3nMpH: 7.4Assay Description:Inhibitory concentration against glutamine synthetase incubated at 37 degree C for 20 minutes in pH 7.4More data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 1.20E+3nMpH: 7.4Assay Description:Inhibitory concentration against glutamine synthetase incubated at 37 degree C for 20 minutes in pH 7.4More data for this Ligand-Target Pair
Affinity DataIC50: 7.60E+3nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 7.60E+3nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 1.04E+4nMpH: 7.4Assay Description:Inhibitory concentration against glutamine synthetase incubated at 37 degree C for 20 minutes in pH 7.4More data for this Ligand-Target Pair
Affinity DataIC50: 1.28E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.28E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.10E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.10E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.12E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.12E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 3.10E+4nMpH: 7.4Assay Description:Inhibitory concentration against glutamine synthetase incubated at 37 degree C for 20 minutes in pH 7.4More data for this Ligand-Target Pair
Affinity DataIC50: 3.19E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.19E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 3.30E+4nMpH: 7.4Assay Description:Inhibitory concentration against glutamine synthetase incubated at 37 degree C for 20 minutes in pH 7.4More data for this Ligand-Target Pair
Affinity DataIC50: 3.30E+4nMAssay Description:Inhibition of Oryza sativa (rice) glutamine synthetaseMore data for this Ligand-Target Pair
Affinity DataIC50: 3.92E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.92E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 5.50E+4nMpH: 7.4Assay Description:Inhibitory concentration against glutamine synthetase incubated at 37 degree C for 20 minutes in pH 7.4More data for this Ligand-Target Pair
Affinity DataIC50: 5.60E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 5.60E+4nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
TargetGlutamine synthetase(Escherichia coli (strain K12))
Wroclaw University Of Technology
Curated by ChEMBL
Wroclaw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 9.70E+5nMpH: 7.4Assay Description:Inhibitory concentration against glutamine synthetase incubated at 37 degree C for 20 minutes in pH 7.4More data for this Ligand-Target Pair
Affinity DataIC50: 1.05E+6nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.05E+6nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.16E+6nMAssay Description:Inhibition of Zea mays cv. Jeff (maize) leaf glutamine synthetase using L-glutamate as substrate assessed as inorganic phosphate production after 20 ...More data for this Ligand-Target Pair