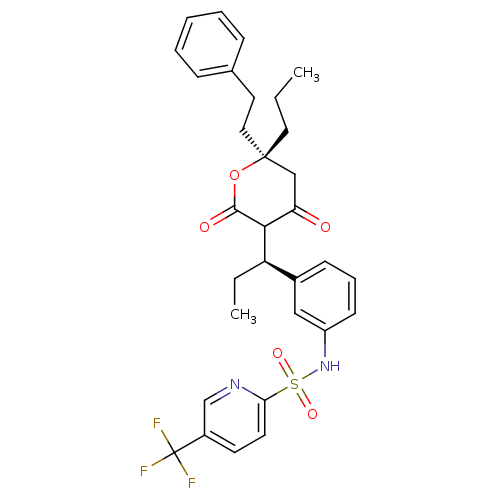
Affinity DataKi: 0.00800nMAssay Description:Inhibition of HIV1 protease by fluorescent peptide substrate based assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.00800nM ΔG°: -62.7kJ/molepH: 5.0 T: 2°CAssay Description:HIV-1 protease was purified and refolded from E. coli inclusion bodies. The substrate used spans the p17-p24 processing site (R-V-S-Q-N-Y-P-I-V-Q-N-K...More data for this Ligand-Target Pair

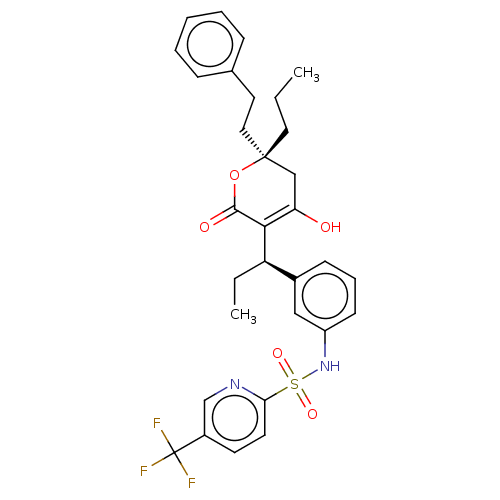
 3D Structure (crystal)
3D Structure (crystal)