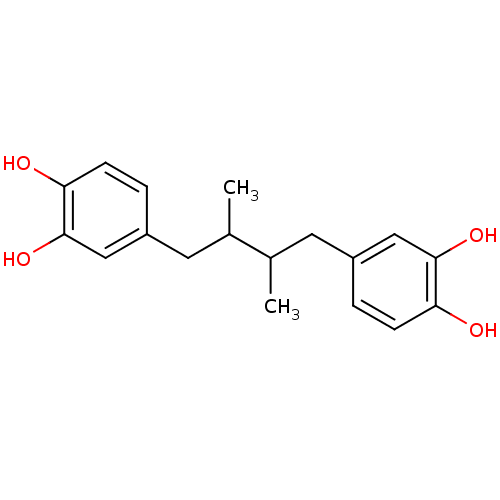
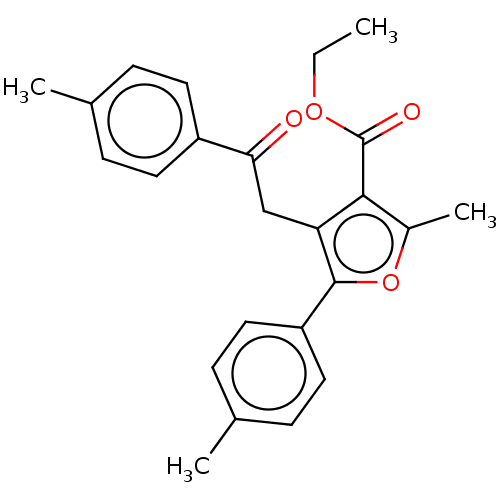
Affinity DataKi: 70.9nMAssay Description:The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus...More data for this Ligand-Target Pair
Affinity DataKi: 141nMAssay Description:The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus...More data for this Ligand-Target Pair
Affinity DataKi: 179nMAssay Description:The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus...More data for this Ligand-Target Pair
Affinity DataKi: 232nMAssay Description:The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus...More data for this Ligand-Target Pair
Affinity DataKi: 246nMAssay Description:The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus...More data for this Ligand-Target Pair
Affinity DataKi: 313nMAssay Description:The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus...More data for this Ligand-Target Pair
Affinity DataKi: 345nMAssay Description:The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus...More data for this Ligand-Target Pair
Affinity DataKi: 357nMAssay Description:The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus...More data for this Ligand-Target Pair
Affinity DataKi: 539nMAssay Description:The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus...More data for this Ligand-Target Pair
Affinity DataKi: 662nMAssay Description:The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus...More data for this Ligand-Target Pair
Affinity DataKi: 868nMAssay Description:The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus...More data for this Ligand-Target Pair
TargetSeed linoleate 13S-lipoxygenase-1(Glycine max (soybean))
Csir-Indian Institute Of Chemical Biology
Csir-Indian Institute Of Chemical Biology
Affinity DataIC50: 1.28E+4nMpH: 8.0Assay Description:SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac...More data for this Ligand-Target Pair
TargetSeed linoleate 13S-lipoxygenase-1(Glycine max (soybean))
Csir-Indian Institute Of Chemical Biology
Csir-Indian Institute Of Chemical Biology
Affinity DataIC50: 2.12E+4nMpH: 8.0Assay Description:SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac...More data for this Ligand-Target Pair
TargetSeed linoleate 13S-lipoxygenase-1(Glycine max (soybean))
Csir-Indian Institute Of Chemical Biology
Csir-Indian Institute Of Chemical Biology
Affinity DataIC50: 2.21E+4nMpH: 8.0Assay Description:SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac...More data for this Ligand-Target Pair
TargetSeed linoleate 13S-lipoxygenase-1(Glycine max (soybean))
Csir-Indian Institute Of Chemical Biology
Csir-Indian Institute Of Chemical Biology
Affinity DataIC50: 2.26E+4nMpH: 8.0Assay Description:SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac...More data for this Ligand-Target Pair
TargetSeed linoleate 13S-lipoxygenase-1(Glycine max (soybean))
Csir-Indian Institute Of Chemical Biology
Csir-Indian Institute Of Chemical Biology
Affinity DataIC50: 3.20E+4nMpH: 8.0Assay Description:SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac...More data for this Ligand-Target Pair
TargetSeed linoleate 13S-lipoxygenase-1(Glycine max (soybean))
Csir-Indian Institute Of Chemical Biology
Csir-Indian Institute Of Chemical Biology
Affinity DataIC50: 3.25E+4nMpH: 8.0Assay Description:SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac...More data for this Ligand-Target Pair
TargetSeed linoleate 13S-lipoxygenase-1(Glycine max (soybean))
Csir-Indian Institute Of Chemical Biology
Csir-Indian Institute Of Chemical Biology
Affinity DataIC50: 3.27E+4nMpH: 8.0Assay Description:SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac...More data for this Ligand-Target Pair
TargetSeed linoleate 13S-lipoxygenase-1(Glycine max (soybean))
Csir-Indian Institute Of Chemical Biology
Csir-Indian Institute Of Chemical Biology
Affinity DataIC50: 4.75E+4nMpH: 8.0Assay Description:SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac...More data for this Ligand-Target Pair
TargetSeed linoleate 13S-lipoxygenase-1(Glycine max (soybean))
Csir-Indian Institute Of Chemical Biology
Csir-Indian Institute Of Chemical Biology
Affinity DataIC50: 5.65E+4nMpH: 8.0Assay Description:SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac...More data for this Ligand-Target Pair
TargetSeed linoleate 13S-lipoxygenase-1(Glycine max (soybean))
Csir-Indian Institute Of Chemical Biology
Csir-Indian Institute Of Chemical Biology
Affinity DataIC50: 7.85E+4nMpH: 8.0Assay Description:SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac...More data for this Ligand-Target Pair
TargetSeed linoleate 13S-lipoxygenase-1(Glycine max (soybean))
Csir-Indian Institute Of Chemical Biology
Csir-Indian Institute Of Chemical Biology
Affinity DataIC50: 2.87E+5nMpH: 8.0Assay Description:SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac...More data for this Ligand-Target Pair