Reaction Details  Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataCell Reactant:
Thymidine Kinase (TK.dT)
Syringe Reactant:
BDBM2
Meas. Tech.:
ITC
Entry Date.:
12/13/00
ΔG°:
-9.04±0.35 (kcal/mole)
pH:
7.5±n/a
Temperature:
298.15±n/a (K)
ΔH° :
-13.87±0.69 (kJ/mole)
Corrected for ΔHioniz:
yes
Protons Released:
-0.31
ΔCp :
-0.14±0.02 (kJ/mole)
ΔS° :
-0.02±0.01 (kJ/mole-K)
Citation
 Perozzo, R; Jelesarov, I; Bosshard, HR; Folkers, G; Scapozza, L Compulsory order of substrate binding to herpes simplex virus type 1 thymidine kinase. A calorimetric study. J Biol Chem 275:16139-45 (2000) [PubMed] Article
Perozzo, R; Jelesarov, I; Bosshard, HR; Folkers, G; Scapozza, L Compulsory order of substrate binding to herpes simplex virus type 1 thymidine kinase. A calorimetric study. J Biol Chem 275:16139-45 (2000) [PubMed] ArticleCell React
Purity:
More than 99% of the enzyme was present as TK.dT complex
Prep. Method:
0.0373 mM enzyme exists in the presence of 1 mM dT.
Name:
Thymidine Kinase (TK.dT)
Synonyms:
thymidine kinase and thymidine complex (TK.dT)
Type:
Protein Complex
Mol. Mass.:
n/a
Description:
n/a
Components:
This complex has 2 components.
Component 1
Name:
Thymidine kinase
Synonyms:
KITH_HHV11 | TK
Type:
Enzyme
Mol. Mass.:
40978.57
Organism:
Human herpesvirus 1
Description:
P03176
Residue:
376
Sequence:
MASYPCHQHASAFDQAARSRGHNNRRTALRPRRQQEATEVRPEQKMPTLLRVYIDGPHGMGKTTTTQLLVALGSRDDIVYVPEPMTYWRVLGASETIANIYTTQHRLDQGEISAGDAAVVMTSAQITMGMPYAVTDAVLAPHIGGEAGSSHAPPPALTLIFDRHPIAALLCYPAARYLMGSMTPQAVLAFVALIPPTLPGTNIVLGALPEDRHIDRLAKRQRPGERLDLAMLAAIRRVYGLLANTVRYLQCGGSWREDWGQLSGTAVPPQGAEPQSNAGPRPHIGDTLFTLFRAPELLAPNGDLYNVFAWALDVLAKRLRSMHVFILDYDQSPAGCRDALLQLTSGMVQTHVTTPGSIPTICDLARTFAREMGEAN
Component 2
Name:
BDBM1
Synonyms:
dT | thymidine
Type:
Nucleoside or nucleotide
Emp. Form.:
C10H14N2O5
Mol. Mass.:
242.2286
SMILES:
Cc1cn([C@H]2C[C@H](O)[C@@H](CO)O2)c(=O)[nH]c1=O
Syringe React
Source:
Sigma
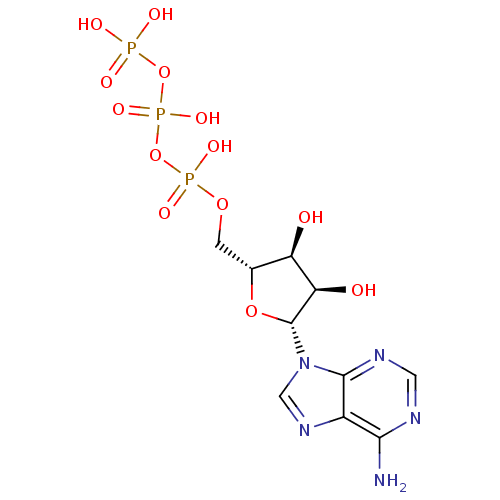
Name:
BDBM2
Synonyms:
({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)phosphonic acid | ATP | Brimonidine ATP | CHEMBL14249
Type:
Nucleoside or nucleotide
Emp. Form.:
C10H16N5O13P3
Mol. Mass.:
507.181
SMILES:
Nc1ncnc2n(cnc12)[C@@H]1O[C@H](COP(O)(=O)OP(O)(=O)OP(O)(O)=O)[C@@H](O)[C@H]1O |r|