Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Acetylcholinesterase
Ligand
BDBM10632
Substrate
BDBM8959
Meas. Tech.
Enzyme Inhibition Assay
pH
7±n/a
Temperature
298.15±n/a K
Ki
334±29 nM
Citation
 Dvir, H; Jiang, HL; Wong, DM; Harel, M; Chetrit, M; He, XC; Jin, GY; Yu, GL; Tang, XC; Silman, I; Bai, DL; Sussman, JL X-ray structures of Torpedo californica acetylcholinesterase complexed with (+)-huperzine A and (-)-huperzine B: structural evidence for an active site rearrangement. Biochemistry 41:10810-8 (2002) [PubMed] Article
Dvir, H; Jiang, HL; Wong, DM; Harel, M; Chetrit, M; He, XC; Jin, GY; Yu, GL; Tang, XC; Silman, I; Bai, DL; Sussman, JL X-ray structures of Torpedo californica acetylcholinesterase complexed with (+)-huperzine A and (-)-huperzine B: structural evidence for an active site rearrangement. Biochemistry 41:10810-8 (2002) [PubMed] Article More Info.:
Target
Name:
Acetylcholinesterase
Synonyms:
3.1.1.7 | ACES_TETCF | Acetylcholinesterase (AChE) | Acetylcholinesterase precursor | ache
Type:
n/a
Mol. Mass.:
65900.74
Organism:
Tetronarce californica (Pacific electric ray) (Torpedo californica)
Description:
P04058
Residue:
586
Sequence:
MNLLVTSSLGVLLHLVVLCQADDHSELLVNTKSGKVMGTRVPVLSSHISAFLGIPFAEPPVGNMRFRRPEPKKPWSGVWNASTYPNNCQQYVDEQFPGFSGSEMWNPNREMSEDCLYLNIWVPSPRPKSTTVMVWIYGGGFYSGSSTLDVYNGKYLAYTEEVVLVSLSYRVGAFGFLALHGSQEAPGNVGLLDQRMALQWVHDNIQFFGGDPKTVTIFGESAGGASVGMHILSPGSRDLFRRAILQSGSPNCPWASVSVAEGRRRAVELGRNLNCNLNSDEELIHCLREKKPQELIDVEWNVLPFDSIFRFSFVPVIDGEFFPTSLESMLNSGNFKKTQILLGVNKDEGSFFLLYGAPGFSKDSESKISREDFMSGVKLSVPHANDLGLDAVTLQYTDWMDDNNGIKNRDGLDDIVGDHNVICPLMHFVNKYTKFGNGTYLYFFNHRASNLVWPEWMGVIHGYEIEFVFGLPLVKELNYTAEEEALSRRIMHYWATFAKTGNPNEPHSQESKWPLFTTKEQKFIDLNTEPMKVHQRLRVQMCVFWNQFLPKLLNATACDGELSSSGTSSSKGIIFYVLFSILYLIF
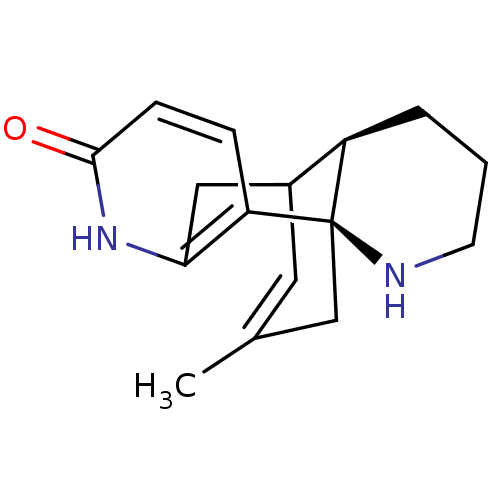
Inhibitor
Name:
BDBM10632
Synonyms:
(-)-Huperzine B | (1R,10R)-16-methyl-6,14-diazatetracyclo[7.5.3.0^{1,10}.0^{2,7}]heptadeca-2(7),3,16-trien-5-one
Type:
Small organic molecule
Emp. Form.:
C16H20N2O
Mol. Mass.:
256.3428
SMILES:
CC1=CC2Cc3[nH]c(=O)ccc3[C@]3(C1)NCCC[C@H]23 |r,t:1,TLB:17:18:4.5.11:2.1.13,THB:10:11:18:2.1.13|
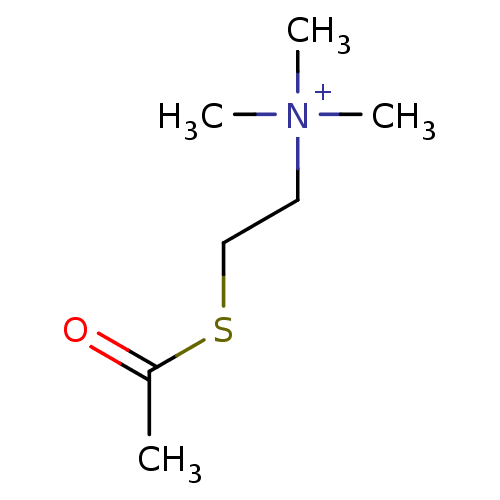
Substrate
Name:
BDBM8959
Synonyms:
(2-Mercaptoethyl)trimethylammonium iodide acetate | ATC | Acetylthiocholine | [2-(acetylsulfanyl)ethyl]trimethylazanium iodide | acetylthiocholine chloride | acetylthiocholine iodide
Type:
Small organic molecule
Emp. Form.:
C7H16NOS
Mol. Mass.:
162.272
SMILES:
CC(=O)SCC[N+](C)(C)C