Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
ATP-dependent molecular chaperone HSP82
Ligand
BDBM15361
Substrate
n/a
Meas. Tech.
FP Assay
IC50
47±n/a nM
Citation
 Proisy, N; Sharp, SY; Boxall, K; Connelly, S; Roe, SM; Prodromou, C; Slawin, AM; Pearl, LH; Workman, P; Moody, CJ Inhibition of Hsp90 with synthetic macrolactones: synthesis and structural and biological evaluation of ring and conformational analogs of radicicol. Chem Biol 13:1203-15 (2006) [PubMed] Article
Proisy, N; Sharp, SY; Boxall, K; Connelly, S; Roe, SM; Prodromou, C; Slawin, AM; Pearl, LH; Workman, P; Moody, CJ Inhibition of Hsp90 with synthetic macrolactones: synthesis and structural and biological evaluation of ring and conformational analogs of radicicol. Chem Biol 13:1203-15 (2006) [PubMed] Article More Info.:
Target
Name:
ATP-dependent molecular chaperone HSP82
Synonyms:
82 kDa heat shock protein | ATP-dependent molecular chaperone HSP82 | HSP82 | HSP82_YEAST | HSP90 | Heat Shock Protein 90 (Hsp90) | Heat shock protein Hsp90 heat-inducible isoform
Type:
Molecular Chaperone
Mol. Mass.:
81369.08
Organism:
Saccharomyces cerevisiae
Description:
Histidine-tagged yeast HSP90 was transformed into E. coli and purified (>90%) by metal affinity, gel filtration, and ion-exchange chromatography.
Residue:
709
Sequence:
MASETFEFQAEITQLMSLIINTVYSNKEIFLRELISNASDALDKIRYKSLSDPKQLETEPDLFIRITPKPEQKVLEIRDSGIGMTKAELINNLGTIAKSGTKAFMEALSAGADVSMIGQFGVGFYSLFLVADRVQVISKSNDDEQYIWESNAGGSFTVTLDEVNERIGRGTILRLFLKDDQLEYLEEKRIKEVIKRHSEFVAYPIQLVVTKEVEKEVPIPEEEKKDEEKKDEEKKDEDDKKPKLEEVDEEEEKKPKTKKVKEEVQEIEELNKTKPLWTRNPSDITQEEYNAFYKSISNDWEDPLYVKHFSVEGQLEFRAILFIPKRAPFDLFESKKKKNNIKLYVRRVFITDEAEDLIPEWLSFVKGVVDSEDLPLNLSREMLQQNKIMKVIRKNIVKKLIEAFNEIAEDSEQFEKFYSAFSKNIKLGVHEDTQNRAALAKLLRYNSTKSVDELTSLTDYVTRMPEHQKNIYYITGESLKAVEKSPFLDALKAKNFEVLFLTDPIDEYAFTQLKEFEGKTLVDITKDFELEETDEEKAEREKEIKEYEPLTKALKEILGDQVEKVVVSYKLLDAPAAIRTGQFGWSANMERIMKAQALRDSSMSSYMSSKKTFEISPKSPIIKELKKRVDEGGAQDKTVKDLTKLLYETALLTSGFSLDEPTSFASRINRLISLGLNIDEDEETETAPEASTAAPVEEVPADTEMEEVD
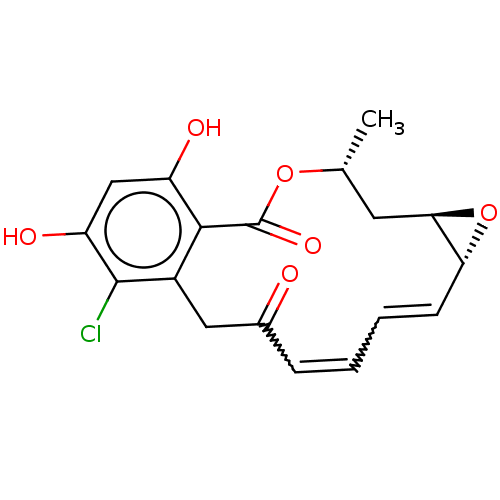
Inhibitor
Name:
BDBM15361
Synonyms:
(1aR,2Z,4E,14R,15aR)-8-chloro-9,11-dihydroxy-14-methyl-1a,14,15,15a-tetrahydro-6H-oxireno[e][2]benzoxacyclotetradecine-6,12(7H)-dione | (4R,6R,8R,9Z,11E)-16-chloro-17,19-dihydroxy-4-methyl-3,7-dioxatricyclo[13.4.0.0^{6,8}]nonadeca-1(19),9,11,15,17-pentaene-2,13-dione | CHEMBL414883 | Microlactone, 1 | Monorden | Radicicol
Type:
Antibiotic
Emp. Form.:
C18H17ClO6
Mol. Mass.:
364.777
SMILES:
[H][C@@]12C[C@@H](C)OC(=O)c3c(O)cc(O)c(Cl)c3CC(=O)C=CC=C[C@@]1([H])O2 |r,w:22.22,20.20|