Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Non-lysosomal glucosylceramidase
Ligand
BDBM50299752
Substrate
n/a
Meas. Tech.
ChEMBL_589172 (CHEMBL1040323)
IC50
>1000000±n/a nM
Citation
 Risseeuw, MD; van den Berg, RJ; Donker-Koopman, WE; van der Marel, GA; Aerts, JM; Overhand, M; Overkleeft, HS Synthesis and evaluation of D-gluco-pyranocyclopropyl amines as potential glucosidase inhibitors. Bioorg Med Chem Lett 19:6600-3 (2009) [PubMed] Article
Risseeuw, MD; van den Berg, RJ; Donker-Koopman, WE; van der Marel, GA; Aerts, JM; Overhand, M; Overkleeft, HS Synthesis and evaluation of D-gluco-pyranocyclopropyl amines as potential glucosidase inhibitors. Bioorg Med Chem Lett 19:6600-3 (2009) [PubMed] Article More Info.:
Target
Name:
Non-lysosomal glucosylceramidase
Synonyms:
Beta-glucosidase | GBA2 | GBA2_HUMAN | KIAA1605 | SPG46
Type:
PROTEIN
Mol. Mass.:
104639.04
Organism:
Homo sapiens (Human)
Description:
ChEMBL_1435476
Residue:
927
Sequence:
MGTQDPGNMGTGVPASEQISCAKEDPQVYCPEETGGTKDVQVTDCKSPEDSRPPKETDCCNPEDSGQLMVSYEGKAMGYQVPPFGWRICLAHEFTEKRKPFQANNVSLSNMIKHIGMGLRYLQWWYRKTHVEKKTPFIDMINSVPLRQIYGCPLGGIGGGTITRGWRGQFCRWQLNPGMYQHRTVIADQFTVCLRREGQTVYQQVLSLERPSVLRSWNWGLCGYFAFYHALYPRAWTVYQLPGQNVTLTCRQITPILPHDYQDSSLPVGVFVWDVENEGDEALDVSIMFSMRNGLGGGDDAPGGLWNEPFCLERSGETVRGLLLHHPTLPNPYTMAVAARVTAATTVTHITAFDPDSTGQQVWQDLLQDGQLDSPTGQSTPTQKGVGIAGAVCVSSKLRPRGQCRLEFSLAWDMPRIMFGAKGQVHYRRYTRFFGQDGDAAPALSHYALCRYAEWEERISAWQSPVLDDRSLPAWYKSALFNELYFLADGGTVWLEVLEDSLPEELGRNMCHLRPTLRDYGRFGYLEGQEYRMYNTYDVHFYASFALIMLWPKLELSLQYDMALATLREDLTRRRYLMSGVMAPVKRRNVIPHDIGDPDDEPWLRVNAYLIHDTADWKDLNLKFVLQVYRDYYLTGDQNFLKDMWPVCLAVMESEMKFDKDHDGLIENGGYADQTYDGWVTTGPSAYCGGLWLAAVAVMVQMAALCGAQDIQDKFSSILSRGQEAYERLLWNGRYYNYDSSSRPQSRSVMSDQCAGQWFLKACGLGEGDTEVFPTQHVVRALQTIFELNVQAFAGGAMGAVNGMQPHGVPDKSSVQSDEVWVGVVYGLAATMIQEGLTWEGFQTAEGCYRTVWERLGLAFQTPEAYCQQRVFRSLAYMRPLSIWAMQLALQQQQHKKASWPKVKQGTGLRTGPMFGPKEAMANLSPE
Inhibitor
Name:
BDBM50299752
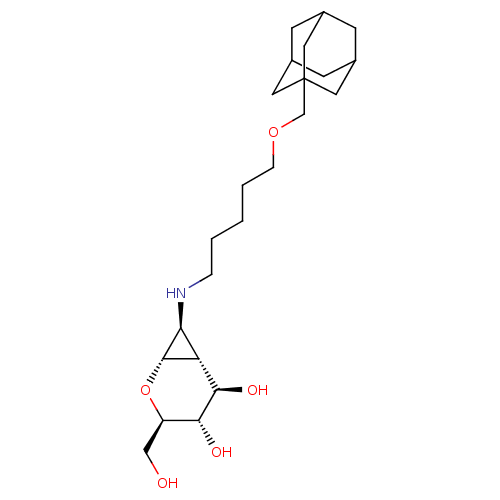
Synonyms:
(1S,3R,4S,5R,6R,7S)-4,5-dihydroxy-3-(hydroxymethyl)-N-(5-(adamant-1-yl)-methoxy pentyl)-2-oxabicyclo[4.1.0]heptan-7-amine | CHEMBL573995
Type:
Small organic molecule
Emp. Form.:
C23H39NO5
Mol. Mass.:
409.5595
SMILES:
OC[C@H]1O[C@@H]2[C@@H](NCCCCCOCC34CC5CC(CC(C5)C3)C4)[C@@H]2[C@@H](O)[C@@H]1O |r,THB:22:14:17:21.20.19,22:20:14.15.23:17,19:18:15:21.20.22,19:20:15:23.17.18,13:14:17:21.20.19|