Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Lysosomal acid glucosylceramidase
Ligand
BDBM18351
Substrate
n/a
Meas. Tech.
ChEMBL_607223 (CHEMBL1070063)
IC50
506000±n/a nM
Citation
 van Scherpenzeel, M; van den Berg, RJ; Donker-Koopman, WE; Liskamp, RM; Aerts, JM; Overkleeft, HS; Pieters, RJ Nanomolar affinity, iminosugar-based chemical probes for specific labeling of lysosomal glucocerebrosidase. Bioorg Med Chem 18:267-73 (2010) [PubMed] Article
van Scherpenzeel, M; van den Berg, RJ; Donker-Koopman, WE; Liskamp, RM; Aerts, JM; Overkleeft, HS; Pieters, RJ Nanomolar affinity, iminosugar-based chemical probes for specific labeling of lysosomal glucocerebrosidase. Bioorg Med Chem 18:267-73 (2010) [PubMed] Article More Info.:
Target
Name:
Lysosomal acid glucosylceramidase
Synonyms:
Acid beta-glucosidase | Alglucerase | Beta-glucocerebrosidase | Beta-glucocerebrosidase (GC) | D-glucosyl-N-acylsphingosine glucohydrolase | GBA | GBA1 | GBA1_HUMAN | GC | GCase | GLUC | Glucocerebrosidase (GBA) | Glucosylceramidase (GBA) | Glucosylceramidase (GCase) | Glucosylceramidase precursor (Beta-glucocerebrosidase) (Acid beta-glucosidase) (D-glucosyl-N-acylsphingosine glucohydrolase) (Alglucerase) (Imiglucerase) | Imiglucerase | beta-glucocerebrosidase (GCase)
Type:
Enzyme
Mol. Mass.:
59724.64
Organism:
Homo sapiens (Human)
Description:
The beta-Glu activity was measured with commercially available beta-glucocerebrosidase (Ceredase) as the enzyme source.
Residue:
536
Sequence:
MEFSSPSREECPKPLSRVSIMAGSLTGLLLLQAVSWASGARPCIPKSFGYSSVVCVCNATYCDSFDPPTFPALGTFSRYESTRSGRRMELSMGPIQANHTGTGLLLTLQPEQKFQKVKGFGGAMTDAAALNILALSPPAQNLLLKSYFSEEGIGYNIIRVPMASCDFSIRTYTYADTPDDFQLHNFSLPEEDTKLKIPLIHRALQLAQRPVSLLASPWTSPTWLKTNGAVNGKGSLKGQPGDIYHQTWARYFVKFLDAYAEHKLQFWAVTAENEPSAGLLSGYPFQCLGFTPEHQRDFIARDLGPTLANSTHHNVRLLMLDDQRLLLPHWAKVVLTDPEAAKYVHGIAVHWYLDFLAPAKATLGETHRLFPNTMLFASEACVGSKFWEQSVRLGSWDRGMQYSHSIITNLLYHVVGWTDWNLALNPEGGPNWVRNFVDSPIIVDITKDTFYKQPMFYHLGHFSKFIPEGSQRVGLVASQKNDLDAVALMHPDGSAVVVVLNRSSKDVPLTIKDPAVGFLETISPGYSIHTYLWRRQ
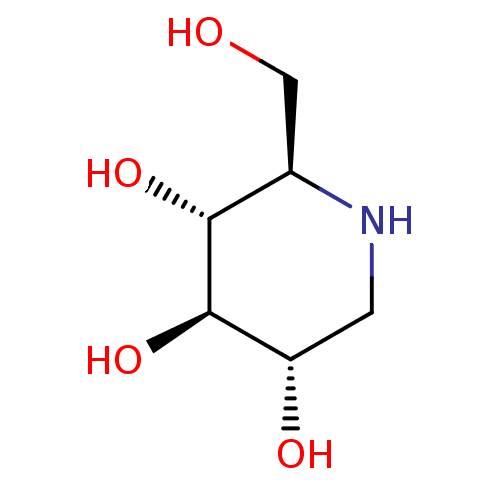
Inhibitor
Name:
BDBM18351
Synonyms:
(2R,3R,4R,5S)-2-(Hydroxymethyl)piperidine-3,4,5-triol, 10 | (2R,3R,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol | 1-Deoxynojirimycin | 1-deoxynojirimycin (DNJ) | CHEMBL307429 | US20230339856, Compound DNJ | US9181184, 1 | dNM
Type:
natural product
Emp. Form.:
C6H13NO4
Mol. Mass.:
163.1717
SMILES:
OC[C@H]1NC[C@H](O)[C@@H](O)[C@@H]1O