Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Substance-P receptor
Ligand
BDBM50002653
Substrate
n/a
Meas. Tech.
ChEBML_208663
IC50
800±n/a nM
Citation
 Howson, W; Hodgson, J; Richardson, R; Walton, L; Guard, S; Watling, K An SAR study for the non-peptide substance P receptor (NK1) antagonist, CP-96,345. Bioorg Med Chem Lett 2:559-564 (1992) Article
Howson, W; Hodgson, J; Richardson, R; Walton, L; Guard, S; Watling, K An SAR study for the non-peptide substance P receptor (NK1) antagonist, CP-96,345. Bioorg Med Chem Lett 2:559-564 (1992) Article More Info.:
Target
Name:
Substance-P receptor
Synonyms:
NK-1 receptor | NK-1R | NK1 Receptor | NK1R_RAT | Neurokinin 1 receptor | Neurokinin NK1 | SPR | Substance-P receptor | Tac1r | Tachykinin receptor 1 | Tacr1
Type:
G Protein-Coupled Receptor (GPCR)
Mol. Mass.:
46371.54
Organism:
Rattus norvegicus (rat)
Description:
Competition binding assays were carried out using membrane preparations from transfected CHO cells that constitutively expressed the rat NK1 receptor.
Residue:
407
Sequence:
MDNVLPMDSDLFPNISTNTSESNQFVQPTWQIVLWAAAYTVIVVTSVVGNVVVIWIILAHKRMRTVTNYFLVNLAFAEACMAAFNTVVNFTYAVHNVWYYGLFYCKFHNFFPIAALFASIYSMTAVAFDRYMAIIHPLQPRLSATATKVVIFVIWVLALLLAFPQGYYSTTETMPSRVVCMIEWPEHPNRTYEKAYHICVTVLIYFLPLLVIGYAYTVVGITLWASEIPGDSSDRYHEQVSAKRKVVKMMIVVVCTFAICWLPFHVFFLLPYINPDLYLKKFIQQVYLASMWLAMSSTMYNPIIYCCLNDRFRLGFKHAFRCCPFISAGDYEGLEMKSTRYLQTQSSVYKVSRLETTISTVVGAHEEEPEEGPKATPSSLDLTSNGSSRSNSKTMTESSSFYSNMLA
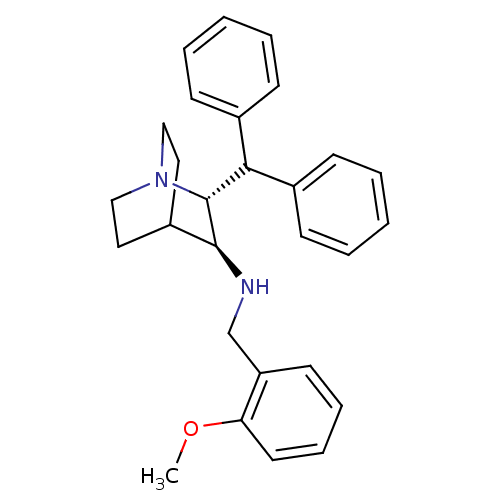
Inhibitor
Name:
BDBM50002653
Synonyms:
((2R,3S)-2-Benzhydryl-1-aza-bicyclo[2.2.2]oct-3-yl)-(2-methoxy-benzyl)-amine | (2-Benzhydryl-1-aza-bicyclo[2.2.2]oct-3-yl)-(2-methoxy-benzyl)-amine | CHEMBL419500 | [(2R,3R)-2-(2,2-Diphenyl-ethyl)-1-aza-bicyclo[2.2.2]oct-3-yl]-(2-methoxy-benzyl)-amine
Type:
Small organic molecule
Emp. Form.:
C28H32N2O
Mol. Mass.:
412.5665
SMILES:
COc1ccccc1CN[C@H]1C2CCN(CC2)[C@@H]1C(c1ccccc1)c1ccccc1 |wU:17.20,wD:10.10,(24.76,-4.86,;23.43,-4.09,;23.45,-2.55,;24.78,-1.78,;24.79,-.24,;23.43,.53,;22.12,-.24,;22.12,-1.78,;20.77,-2.54,;20.77,-4.09,;19.44,-4.85,;18.11,-4.08,;16.78,-4.85,;16.78,-6.39,;18.11,-7.16,;17.53,-6.17,;18.88,-5.41,;19.44,-6.39,;20.77,-7.17,;20.77,-8.71,;22.1,-9.47,;22.1,-11.01,;20.77,-11.78,;19.44,-11.01,;19.44,-9.47,;22.31,-7.16,;23.08,-8.5,;24.62,-8.5,;25.39,-7.17,;24.62,-5.83,;23.08,-5.84,)|