Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
cAMP-dependent protein kinase catalytic subunit alpha
Ligand
BDBM24773
Substrate
n/a
Meas. Tech.
ChEMBL_587098 (CHEMBL1051309)
Kd
>10000±n/a nM
Citation
 Karaman, MW; Herrgard, S; Treiber, DK; Gallant, P; Atteridge, CE; Campbell, BT; Chan, KW; Ciceri, P; Davis, MI; Edeen, PT; Faraoni, R; Floyd, M; Hunt, JP; Lockhart, DJ; Milanov, ZV; Morrison, MJ; Pallares, G; Patel, HK; Pritchard, S; Wodicka, LM; Zarrinkar, PP A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol 26:127-32 (2008) [PubMed] Article
Karaman, MW; Herrgard, S; Treiber, DK; Gallant, P; Atteridge, CE; Campbell, BT; Chan, KW; Ciceri, P; Davis, MI; Edeen, PT; Faraoni, R; Floyd, M; Hunt, JP; Lockhart, DJ; Milanov, ZV; Morrison, MJ; Pallares, G; Patel, HK; Pritchard, S; Wodicka, LM; Zarrinkar, PP A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol 26:127-32 (2008) [PubMed] Article More Info.:
Target
Name:
cAMP-dependent protein kinase catalytic subunit alpha
Synonyms:
KAPCA_HUMAN | PKA C-alpha | PKACA | PRKACA | cAMP-dependent protein kinase (PKA) | cAMP-dependent protein kinase catalytic (PKA) | cAMP-dependent protein kinase catalytic subunit alpha | cAMP-dependent protein kinase catalytic subunit alpha (PKA) | cAMP-dependent protein kinase catalytic subunit alpha (PKACA) | cAMP-dependent protein kinase catalytic subunit alpha (PKAc) | cAMP-dependent protein kinase catalytic subunit alpha isoform 1 | cAMP-dependent protein kinase, alpha-catalytic subunit
Type:
Enzyme Catalytic Subunit
Mol. Mass.:
40598.73
Organism:
Homo sapiens (Human)
Description:
n/a
Residue:
351
Sequence:
MGNAAAAKKGSEQESVKEFLAKAKEDFLKKWESPAQNTAHLDQFERIKTLGTGSFGRVMLVKHKETGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVSINEKCGKEFSEF
Inhibitor
Name:
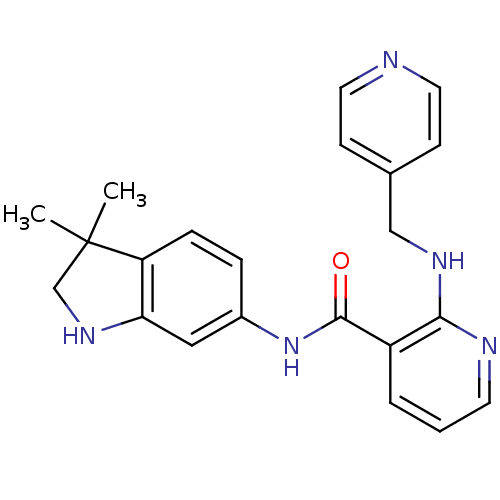
BDBM24773
Synonyms:
AMG 706 | AMG-706 | Motesanib | N-(3,3-dimethyl-1,2-dihydroindol-6-yl)-2-(pyridin-4-ylmethylamino)-3-pyridinecarboxamide | N-(3,3-dimethyl-1,2-dihydroindol-6-yl)-2-(pyridin-4-ylmethylamino)pyridine-3-carboxamide | N-(3,3-dimethyl-2,3-dihydro-1H-indol-6-yl)-2-[(pyridin-4-ylmethyl)amino]pyridine-3-carboxamide | N-(3,3-dimethylindolin-6-yl)-2-(4-pyridylmethylamino)nicotinamide | US10464902, Motesanib | cid_11667893
Type:
Small organic molecule
Emp. Form.:
C22H23N5O
Mol. Mass.:
373.4509
SMILES:
CC1(C)CNc2cc(NC(=O)c3cccnc3NCc3ccncc3)ccc12