Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Potassium voltage-gated channel subfamily H member 2
Ligand
BDBM50131427
Substrate
n/a
Meas. Tech.
ChEMBL_725152 (CHEMBL1676103)
IC50
12022.64±n/a nM
Citation
More Info.:
Target
Name:
Potassium voltage-gated channel subfamily H member 2
Synonyms:
1,3-beta-glucan synthase component GLS2 | Cytochrome P450 3A4 | ERG | ERG1 | Eag-related protein 1 | Ether a-go-go related gene potassium channel (hERG) | Ether-a-go-go-related gene (HERG) | Ether-a-go-go-related gene potassium channel (hERG) | Ether-a-go-go-related gene potassium channel 1 | Ether-a-go-go-related gene potassium channel 1 (HERG) | Ether-a-go-go-related gene potassium channel 1 (hERG1) | Ether-a-go-go-related protein (hERG) | Ether-a-go-go-related protein 1 | Ether-a-go-go-related protein 1 (HERG) | H-ERG | HERG | KCNH2 | KCNH2_HUMAN | Potassium voltage-gated channel subfamily H member 2 (hERG) | Transcriptional regulator ERG | Voltage-gated potassium channel subunit Kv11.1 | eag homolog | hERG Potassium Channel 1 | putative potassium channel subunit
Type:
Multi-pass membrane protein
Mol. Mass.:
126672.65
Organism:
Homo sapiens (Human)
Description:
Q12809
Residue:
1159
Sequence:
MPVRRGHVAPQNTFLDTIIRKFEGQSRKFIIANARVENCAVIYCNDGFCELCGYSRAEVMQRPCTCDFLHGPRTQRRAAAQIAQALLGAEERKVEIAFYRKDGSCFLCLVDVVPVKNEDGAVIMFILNFEVVMEKDMVGSPAHDTNHRGPPTSWLAPGRAKTFRLKLPALLALTARESSVRSGGAGGAGAPGAVVVDVDLTPAAPSSESLALDEVTAMDNHVAGLGPAEERRALVGPGSPPRSAPGQLPSPRAHSLNPDASGSSCSLARTRSRESCASVRRASSADDIEAMRAGVLPPPPRHASTGAMHPLRSGLLNSTSDSDLVRYRTISKIPQITLNFVDLKGDPFLASPTSDREIIAPKIKERTHNVTEKVTQVLSLGADVLPEYKLQAPRIHRWTILHYSPFKAVWDWLILLLVIYTAVFTPYSAAFLLKETEEGPPATECGYACQPLAVVDLIVDIMFIVDILINFRTTYVNANEEVVSHPGRIAVHYFKGWFLIDMVAAIPFDLLIFGSGSEELIGLLKTARLLRLVRVARKLDRYSEYGAAVLFLLMCTFALIAHWLACIWYAIGNMEQPHMDSRIGWLHNLGDQIGKPYNSSGLGGPSIKDKYVTALYFTFSSLTSVGFGNVSPNTNSEKIFSICVMLIGSLMYASIFGNVSAIIQRLYSGTARYHTQMLRVREFIRFHQIPNPLRQRLEEYFQHAWSYTNGIDMNAVLKGFPECLQADICLHLNRSLLQHCKPFRGATKGCLRALAMKFKTTHAPPGDTLVHAGDLLTALYFISRGSIEILRGDVVVAILGKNDIFGEPLNLYARPGKSNGDVRALTYCDLHKIHRDDLLEVLDMYPEFSDHFWSSLEITFNLRDTNMIPGSPGSTELEGGFSRQRKRKLSFRRRTDKDTEQPGEVSALGPGRAGAGPSSRGRPGGPWGESPSSGPSSPESSEDEGPGRSSSPLRLVPFSSPRPPGEPPGGEPLMEDCEKSSDTCNPLSGAFSGVSNIFSFWGDSRGRQYQELPRCPAPTPSLLNIPLSSPGRRPRGDVESRLDALQRQLNRLETRLSADMATVLQLLQRQMTLVPPAYSAVTTPGPGPTSTSPLLPVSPLPTLTLDSLSQVSQFMACEELPPGAPELPQEGPTRRLSLPGQLGALTSQPLHRHGSDPGS
Inhibitor
Name:
BDBM50131427
Synonyms:
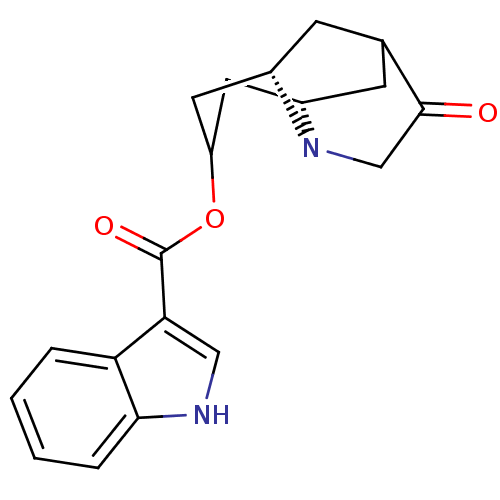
1H-Indole-3-carboxylic acid (3S,7S)-10-oxo-8-aza-tricyclo[5.3.1.0*3,8*]undec-5-yl ester
Type:
Small organic molecule
Emp. Form.:
C19H20N2O3
Mol. Mass.:
324.3737
SMILES:
O=C(OC1C[C@@H]2CC3C[C@@H](C1)N2CC3=O)c1c[nH]c2ccccc12 |wD:9.11,5.12,TLB:14:13:5.6:9.8,12:11:3.4.10:7.6.8,(-1.14,-4.4,;-1.14,-2.86,;.19,-2.08,;1.54,-2.85,;1.54,-4.4,;2.86,-5.15,;2.86,-3.61,;6.86,-2.86,;5.53,-2.08,;4.19,-2.85,;2.86,-2.07,;4.19,-4.4,;5.53,-5.17,;6.86,-4.4,;8.19,-5.18,;-2.47,-2.09,;-2.63,-.55,;-4.15,-.23,;-4.9,-1.57,;-6.41,-1.88,;-6.89,-3.35,;-5.85,-4.5,;-4.35,-4.17,;-3.87,-2.71,)|