Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
G-protein coupled bile acid receptor 1
Ligand
BDBM50003573
Substrate
n/a
Meas. Tech.
ChEMBL_1335566 (CHEMBL3239421)
EC50
1.000000±n/a nM
Citation
 Phillips, DP; Gao, W; Yang, Y; Zhang, G; Lerario, IK; Lau, TL; Jiang, J; Wang, X; Nguyen, DG; Bhat, BG; Trotter, C; Sullivan, H; Welzel, G; Landry, J; Chen, Y; Joseph, SB; Li, C; Gordon, WP; Richmond, W; Johnson, K; Bretz, A; Bursulaya, B; Pan, S; McNamara, P; Seidel, HM Discovery of trifluoromethyl(pyrimidin-2-yl)azetidine-2-carboxamides as potent, orally bioavailable TGR5 (GPBAR1) agonists: structure-activity relationships, lead optimization, and chronic in vivo efficacy. J Med Chem 57:3263-82 (2014) [PubMed] Article
Phillips, DP; Gao, W; Yang, Y; Zhang, G; Lerario, IK; Lau, TL; Jiang, J; Wang, X; Nguyen, DG; Bhat, BG; Trotter, C; Sullivan, H; Welzel, G; Landry, J; Chen, Y; Joseph, SB; Li, C; Gordon, WP; Richmond, W; Johnson, K; Bretz, A; Bursulaya, B; Pan, S; McNamara, P; Seidel, HM Discovery of trifluoromethyl(pyrimidin-2-yl)azetidine-2-carboxamides as potent, orally bioavailable TGR5 (GPBAR1) agonists: structure-activity relationships, lead optimization, and chronic in vivo efficacy. J Med Chem 57:3263-82 (2014) [PubMed] Article More Info.:
Target
Name:
G-protein coupled bile acid receptor 1
Synonyms:
GPBAR_MOUSE | Gpbar1 | M-BAR | Membrane-type receptor for bile acids | Tgr5
Type:
PROTEIN
Mol. Mass.:
35665.78
Organism:
Mus musculus
Description:
ChEMBL_1475560
Residue:
329
Sequence:
MMTPNSTELSAIPMGVLGLSLALASLIVIANLLLALGIALDRHLRSPPAGCFFLSLLLAGLLTGLALPMLPGLWSRNHQGYWSCLLLHLTPNFCFLSLLANLLLVHGERYMAVLQPLRPHGSVRLALFLTWVSSLFFASLPALGWNHWSPDANCSSQAVFPAPYLYLEVYGLLLPAVGATALLSVRVLATAHRQLCEIRRLERAVCRDVPSTLARALTWRQARAQAGATLLFLLCWGPYVATLLLSVLAYERRPPLGPGTLLSLISLGSTSAAAVPVAMGLGDQRYTAPWRTAAQRCLRVLRGRAKRDNPGPSTAYHTSSQCSIDLDLN
Inhibitor
Name:
BDBM50003573
Synonyms:
CHEMBL3234871
Type:
Small organic molecule
Emp. Form.:
C31H30F3N5O3
Mol. Mass.:
577.5968
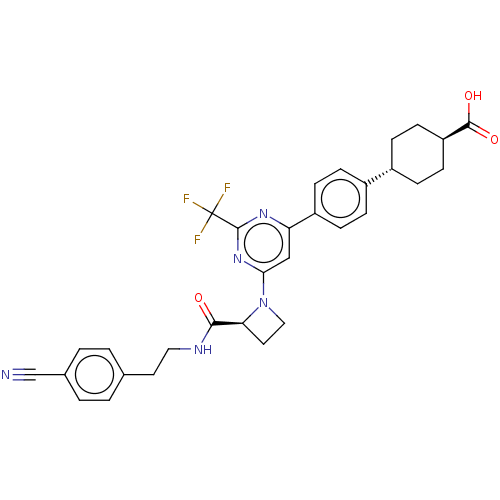
SMILES:
OC(=O)[C@H]1CC[C@@H](CC1)c1ccc(cc1)-c1cc(nc(n1)C(F)(F)F)N1CC[C@H]1C(=O)NCCc1ccc(cc1)C#N |r,wU:6.9,28.32,wD:3.2,(43.64,-56.93,;44.97,-56.16,;46.31,-56.93,;44.97,-54.62,;46.31,-53.85,;46.3,-52.3,;44.97,-51.54,;43.65,-52.32,;43.64,-53.85,;44.96,-50,;43.62,-49.23,;43.62,-47.69,;44.96,-46.92,;46.29,-47.69,;46.29,-49.23,;44.96,-45.38,;46.29,-44.61,;46.29,-43.07,;44.96,-42.3,;43.63,-43.07,;43.62,-44.61,;42.29,-42.3,;40.95,-43.07,;42.29,-40.76,;40.95,-41.53,;47.63,-42.3,;48.02,-40.81,;49.51,-41.22,;49.11,-42.7,;49.87,-44.04,;49.1,-45.37,;51.41,-44.04,;52.19,-42.71,;53.73,-42.72,;54.5,-41.38,;56.04,-41.4,;56.81,-40.07,;56.04,-38.73,;54.49,-38.73,;53.73,-40.06,;56.81,-37.4,;57.58,-36.07,)|