Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
DNA helicase [35-727]
Ligand
BDBM50055660
Substrate
n/a
Meas. Tech.
DNA-Dependent ATPase Assay
pH
8±0
Temperature
310.15±0 K
IC50
3.1e+3±n/a nM
Citation
 Tarique, M; Tabassum, F; Ahmad, M; Tuteja, R Plasmodium falciparum UvrD activities are downregulated by DNA-interacting compounds and its dsRNA inhibits malaria parasite growth. BMC Biochem 15:9 (2014) [PubMed] Article
Tarique, M; Tabassum, F; Ahmad, M; Tuteja, R Plasmodium falciparum UvrD activities are downregulated by DNA-interacting compounds and its dsRNA inhibits malaria parasite growth. BMC Biochem 15:9 (2014) [PubMed] Article More Info.:
Target
Name:
DNA helicase [35-727]
Synonyms:
UvrD helicase (PfUDN)
Type:
Enzyme
Mol. Mass.:
81959.48
Organism:
Plasmodium falciparum
Description:
N-terminal fragment of P. falciparum UvrD helicase (35-727 aa)
Residue:
693
Sequence:
NFSEEQQRIIEIPMNVNLCIIACPGSGKTSTLTARIIKSIIEEKQSIVCITFTNYAASDLKDKIMKKINCLIDICVDNKINQKLFNNKNNKINFSLKNKCTLNNKMNKSIFKVLNTVMFIGTIHSFCRYILYKYKGTFKILTDFINTNIIKLAFNNFYSSMMSKTKGTQPGFSTILERKSNKASTQNCDPDKINTHNNDDNINNKNDYINNKNKNDYNNINNYDNINNYDNINNDDNINNDDNINNDDNINNDDNINNDDDINNCGNCNQPKGIPSQLAYFINCMKNAEIKEDEEKEFYEEEHDIQNDILNNDDNNNDEDDDDDDEFYNYLYNFKHSYEQTNDYFANEQVQSVLKKKNIIFLKKKIKLMKYIELYNIKIEINDVEKMFYEEYKKIFKKAKNIYYDFDDLLIETYRLMKDNADIRNKILEEWNYVFCDEFQDTNTTQFNILQFFVNHNVPSTLDQSIYTSGNLENKDIKDNHNISMSTYKSKQYSHSVQNFFNEHKVDQQLCDQIYFDKHCNNILMQNKNQQATHKHSEEEEEQEEEKKKNKKTKKTNTFINQQKIKTQTVLSYKQDEMSSSASSTYSYVKIEKEKKKYALNKIDDTYYNKKNIYSNNDEILDIENEQTFFNNNCNEKNKTKKKCNLKDRSLTVIGDDDQSIYSFRGAHINVFYKFLKDCNCLLFKLNNNFRST
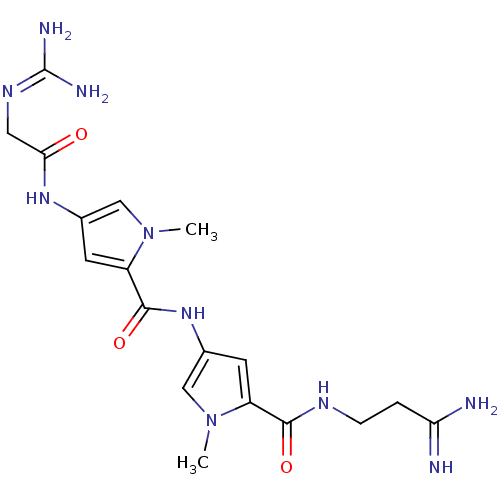
Inhibitor
Name:
BDBM50055660
Synonyms:
1-amino-3-(4-(4-(2-(amino(iminio)methylamino)acetamido)-1-methyl-1H-pyrrole-2-carboxamido)-1-methyl-1H-pyrrole-2-carboxamido)propan-1-iminium | 2N-(3-amino-3-iminopropyl)-4-[4-amino(imino)methylaminomethylcarboxamido-1-methyl-1H-2-pyrrolylcarboxamido]-1-methyl-1H-2-pyrrolecarboxamide | 2N-(3-amino-3-iminopropyl)-4-[4-amino(imino)methylaminomethylcarboxamido-1-methyl-1H-2-pyrrolylcarboxamido]-1-methyl-1H-2-pyrrolecarboxamide(netropsin) | 4-(2-Guanidino-acetylamino)-1-methyl-1H-pyrrole-2-carboxylic acid [5-(2-carbamimidoyl-ethylcarbamoyl)-1-methyl-2,3-dihydro-1H-pyrrol-3-yl]-amide | CHEMBL307767 | NETROPSIN
Type:
Small organic molecule
Emp. Form.:
C18H26N10O3
Mol. Mass.:
430.4642
SMILES:
[#6]-n1cc(-[#7]-[#6](=O)-c2cc(-[#7]-[#6](=O)-[#6]\[#7]=[#6](\[#7])-[#7])cn2-[#6])cc1-[#6](=O)-[#7]-[#6]-[#6]-[#6](-[#7])=[#7]