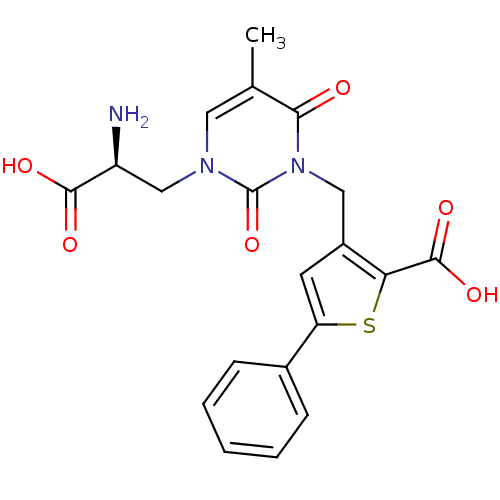
BDBM50207591 (S)-1-(2-amino-2-carboxyethyl)-3-(2-carboxy-5-phenylthiophene-3-yl-methyl)-5-methylpyrimidine-2,4-dione::3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-5-phenylthiophene-2-carboxylic acid::CHEMBL373429
SMILES Cc1cn(C[C@H](N)C(O)=O)c(=O)n(Cc2cc(sc2C(O)=O)-c2ccccc2)c1=O
InChI Key InChIKey=LCZDCKMQSBGXAH-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 50207591
Found 3 hits for monomerid = 50207591
Affinity DataKd: 12nMAssay Description:Activity at native GLUK5 kainate receptor assessed as antagonism of kainite-induced depolarization of neonatal anaesthetised rat dorsal root fibresMore data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+5nMAssay Description:Antagonist activity at human recombinant GLUK6 expressed in HEK293 cells assessed as inhibition of glutamate-stimulated calcium influx by FLIPR assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+5nMAssay Description:Antagonist activity at human recombinant GLUA2-AMPA receptor expressed in HEK293 cells assessed as inhibition of glutamate-stimulated calcium influx ...More data for this Ligand-Target Pair