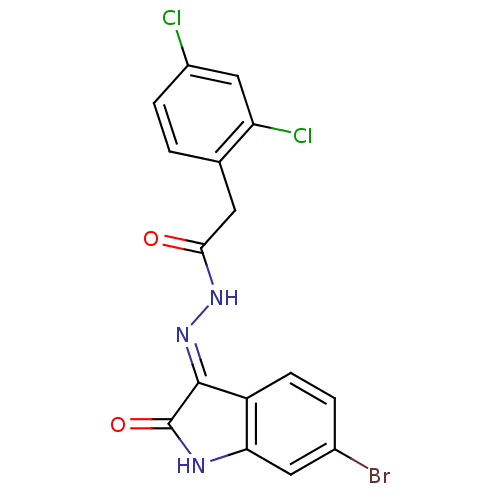
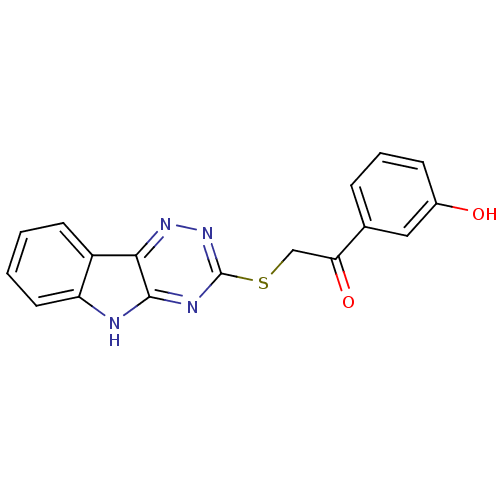
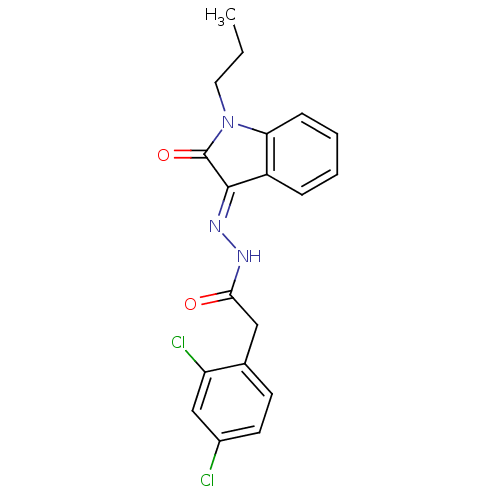
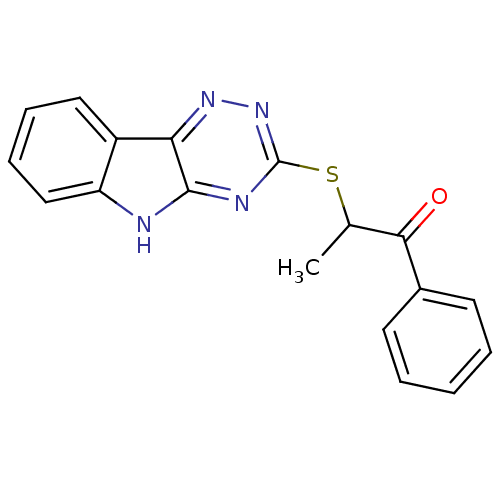
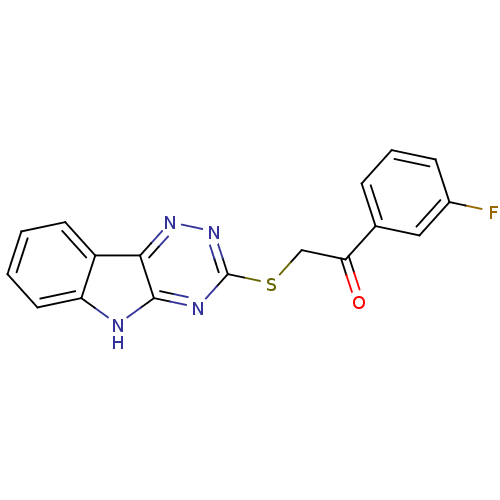
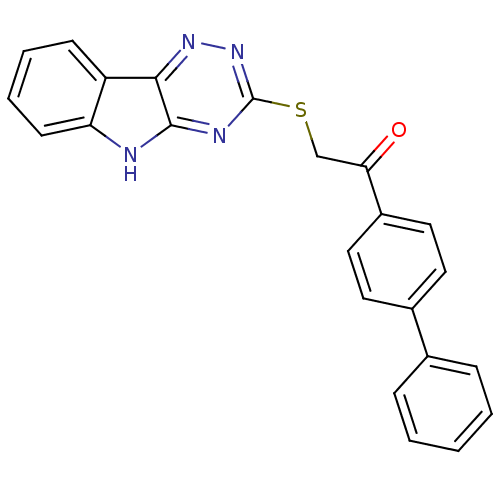
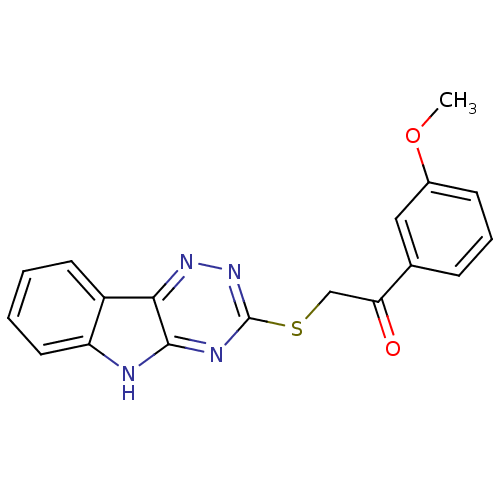
Affinity DataIC50: 2.20E+3nMAssay Description:10 無 of test samples (5 mg/mL DMSO solution) were reconstituted in 100 無 of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
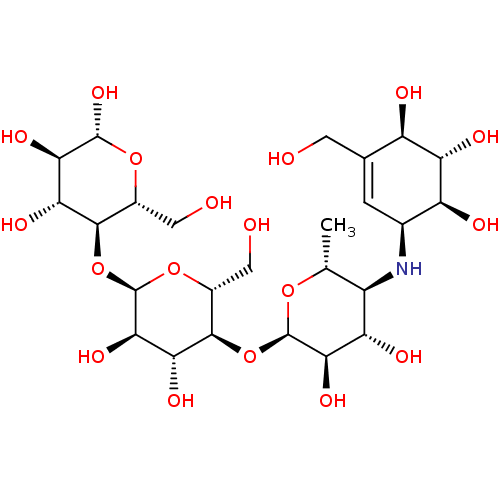
Affinity DataIC50: 2.46E+3nMT: 2°CAssay Description:Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat...More data for this Ligand-Target Pair
Affinity DataIC50: 3.00E+3nMAssay Description:10 無 of test samples (5 mg/mL DMSO solution) were reconstituted in 100 無 of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 3.30E+3nMAssay Description:10 無 of test samples (5 mg/mL DMSO solution) were reconstituted in 100 無 of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 1.18E+4nMAssay Description:10 無 of test samples (5 mg/mL DMSO solution) were reconstituted in 100 無 of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 1.83E+4nMAssay Description:10 無 of test samples (5 mg/mL DMSO solution) were reconstituted in 100 無 of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 2.89E+4nMT: 2°CAssay Description:Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat...More data for this Ligand-Target Pair
Affinity DataIC50: 3.69E+4nMT: 2°CAssay Description:Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat...More data for this Ligand-Target Pair
Affinity DataIC50: 3.71E+4nMT: 2°CAssay Description:Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat...More data for this Ligand-Target Pair
Affinity DataIC50: 3.74E+4nMT: 2°CAssay Description:Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat...More data for this Ligand-Target Pair
Affinity DataIC50: 3.78E+4nMT: 2°CAssay Description:Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat...More data for this Ligand-Target Pair
Affinity DataIC50: 3.81E+4nMT: 2°CAssay Description:Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat...More data for this Ligand-Target Pair
Affinity DataIC50: 3.83E+4nMpH: 6.8Assay Description:Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat...More data for this Ligand-Target Pair
Affinity DataIC50: 5.16E+4nMT: 2°CAssay Description:Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat...More data for this Ligand-Target Pair
Affinity DataIC50: 7.84E+4nMT: 2°CAssay Description:Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat...More data for this Ligand-Target Pair
Affinity DataIC50: 8.35E+4nMAssay Description:10 無 of test samples (5 mg/mL DMSO solution) were reconstituted in 100 無 of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 1.25E+5nMT: 2°CAssay Description:Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat...More data for this Ligand-Target Pair
Affinity DataIC50: 3.13E+5nMT: 2°CAssay Description:Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat...More data for this Ligand-Target Pair
Affinity DataIC50: 8.40E+5nMAssay Description:10 無 of test samples (5 mg/mL DMSO solution) were reconstituted in 100 無 of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair