Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Tyrosine-protein phosphatase non-receptor type 1 [1-298]
Ligand
BDBM13467
Substrate
BDBM13466
Meas. Tech.
Phosphatase Inhibition Assay
pH
7±n/a
Temperature
295.15±n/a K
IC50
0.4±n/a nM
Citation
 Ala, PJ; Gonneville, L; Hillman, M; Becker-Pasha, M; Yue, EW; Douty, B; Wayland, B; Polam, P; Crawley, ML; McLaughlin, E; Sparks, RB; Glass, B; Takvorian, A; Combs, AP; Burn, TC; Hollis, GF; Wynn, R Structural insights into the design of nonpeptidic isothiazolidinone-containing inhibitors of protein-tyrosine phosphatase 1B. J Biol Chem 281:38013-21 (2006) [PubMed] Article
Ala, PJ; Gonneville, L; Hillman, M; Becker-Pasha, M; Yue, EW; Douty, B; Wayland, B; Polam, P; Crawley, ML; McLaughlin, E; Sparks, RB; Glass, B; Takvorian, A; Combs, AP; Burn, TC; Hollis, GF; Wynn, R Structural insights into the design of nonpeptidic isothiazolidinone-containing inhibitors of protein-tyrosine phosphatase 1B. J Biol Chem 281:38013-21 (2006) [PubMed] Article Target
Name:
Tyrosine-protein phosphatase non-receptor type 1 [1-298]
Synonyms:
PTN1_HUMAN | PTP-1B | PTP1B | PTPN1 | PTPase 1B | Protein-Tyrosine Phosphatase 1B (PTP1B) | Tyrosine-protein phosphatase, non-receptor type 1
Type:
Enzyme
Mol. Mass.:
34670.65
Organism:
Homo sapiens (Human)
Description:
The catalytic domain of PTP 1B (residues 1-298) was expressed and purified from E. coli.
Residue:
298
Sequence:
MEMEKEFEQIDKSGSWAAIYQDIRHEASDFPCRVAKLPKNKNRNRYRDVSPFDHSRIKLHQEDNDYINASLIKMEEAQRSYILTQGPLPNTCGHFWEMVWEQKSRGVVMLNRVMEKGSLKCAQYWPQKEEKEMIFEDTNLKLTLISEDIKSYYTVRQLELENLTTQETREILHFHYTTWPDFGVPESPASFLNFLFKVRESGSLSPEHGPVVVHCSAGIGRSGTFCLADTCLLLMDKRKDPSSVDIKKVLLEMRKFRMGLIQTADQLRFSYLAVIEGAKFIMGDSSVQDQWKELSHED
Inhibitor
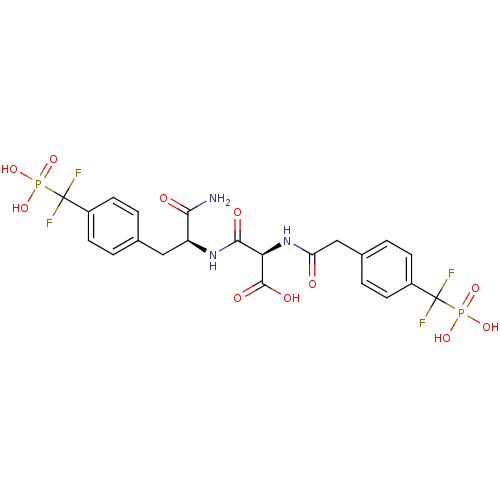
Name:
BDBM13467
Synonyms:
(2R)-2-{[(1S)-1-carbamoyl-2-{4-[difluoro(phosphono)methyl]phenyl}ethyl]carbamoyl}-2-(1-{4-[difluoro(phosphono)methyl]phenyl}acetamido)acetic acid | Difluoromethylphosphonic acid (DFMP) deriv. 7
Type:
Small organic molecule
Emp. Form.:
C22H23F4N3O11P2
Mol. Mass.:
643.3727
SMILES:
NC(=O)[C@H](Cc1ccc(cc1)C(F)(F)P(O)(O)=O)NC(=O)[C@@H](NC(=O)Cc1ccc(cc1)C(F)(F)P(O)(O)=O)C(O)=O |r|
Substrate
Name:
BDBM13466
Synonyms:
4-Nitrophenyl phosphate disodium salt hexahydrate | 4-nitrophenyl phosphate (pNPP) | disodium (4-nitrophenyl) phosphate | para-nitrophenyl phosphate (pNPP)
Type:
Small organic molecule
Emp. Form.:
C6H4NO6P
Mol. Mass.:
217.0739
SMILES:
[O-][N+](=O)c1ccc(O[P+]([O-])([O-])[O-])cc1