Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Matrix metalloproteinase-9
Ligand
BDBM50530230
Substrate
n/a
Meas. Tech.
ChEMBL_1911527 (CHEMBL4413973)
Ki
2588±n/a nM
Citation
 Covaleda, G; Gallego, P; Vendrell, J; Georgiadis, D; Lorenzo, J; Dive, V; Aviles, FX; Reverter, D; Devel, L Synthesis and Structural/Functional Characterization of Selective M14 Metallocarboxypeptidase Inhibitors Based on Phosphinic Pseudopeptide Scaffold: Implications on the Design of Specific Optical Probes. J Med Chem 62:1917-1931 (2019) [PubMed] Article
Covaleda, G; Gallego, P; Vendrell, J; Georgiadis, D; Lorenzo, J; Dive, V; Aviles, FX; Reverter, D; Devel, L Synthesis and Structural/Functional Characterization of Selective M14 Metallocarboxypeptidase Inhibitors Based on Phosphinic Pseudopeptide Scaffold: Implications on the Design of Specific Optical Probes. J Med Chem 62:1917-1931 (2019) [PubMed] Article More Info.:
Target
Name:
Matrix metalloproteinase-9
Synonyms:
67 kDa matrix metalloproteinase-9 | 82 kDa matrix metalloproteinase-9 | 92 kDa gelatinase | 92 kDa type IV collagenase | CLG4B | GELB | Gelatinase B | MMP-9 | MMP9 | MMP9_HUMAN | Matrix metalloproteinase 9 (MMP-9) | Matrix metalloproteinase-9 (MMP-9) | Matrix metalloproteinase-9 (MMP9)
Type:
Enzyme
Mol. Mass.:
78452.28
Organism:
Homo sapiens (Human)
Description:
P14780
Residue:
707
Sequence:
MSLWQPLVLVLLVLGCCFAAPRQRQSTLVLFPGDLRTNLTDRQLAEEYLYRYGYTRVAEMRGESKSLGPALLLLQKQLSLPETGELDSATLKAMRTPRCGVPDLGRFQTFEGDLKWHHHNITYWIQNYSEDLPRAVIDDAFARAFALWSAVTPLTFTRVYSRDADIVIQFGVAEHGDGYPFDGKDGLLAHAFPPGPGIQGDAHFDDDELWSLGKGVVVPTRFGNADGAACHFPFIFEGRSYSACTTDGRSDGLPWCSTTANYDTDDRFGFCPSERLYTQDGNADGKPCQFPFIFQGQSYSACTTDGRSDGYRWCATTANYDRDKLFGFCPTRADSTVMGGNSAGELCVFPFTFLGKEYSTCTSEGRGDGRLWCATTSNFDSDKKWGFCPDQGYSLFLVAAHEFGHALGLDHSSVPEALMYPMYRFTEGPPLHKDDVNGIRHLYGPRPEPEPRPPTTTTPQPTAPPTVCPTGPPTVHPSERPTAGPTGPPSAGPTGPPTAGPSTATTVPLSPVDDACNVNIFDAIAEIGNQLYLFKDGKYWRFSEGRGSRPQGPFLIADKWPALPRKLDSVFEERLSKKLFFFSGRQVWVYTGASVLGPRRLDKLGLGADVAQVTGALRSGRGKMLLFSGRRLWRFDVKAQMVDPRSASEVDRMFPGVPLDTHDVFQYREKAYFCQDRFYWRVSSRSELNQVDQVGYVTYDILQCPED
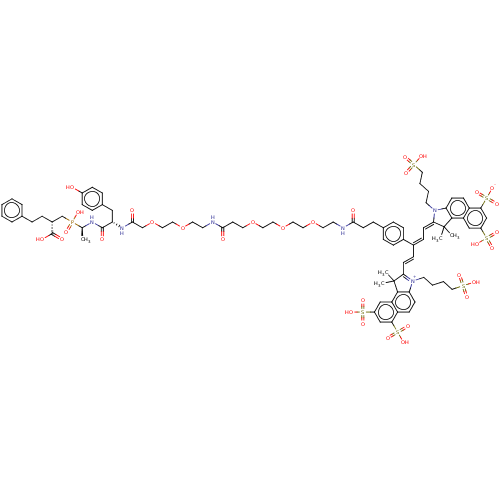
Inhibitor
Name:
BDBM50530230
Synonyms:
CHEMBL4445882
Type:
Small organic molecule
Emp. Form.:
C87H109N6O32PS6
Mol. Mass.:
1974.181
SMILES:
C[C@H](NC(=O)[C@H](Cc1ccc(O)cc1)NC(=O)COCCOCCNC(=O)CCOCCOCCOCCNC(=O)CCc1ccc(cc1)\C(\C=C\C1=[N+](CCCCS(O)(=O)=O)c2ccc3c(cc(cc3c2C1(C)C)S(O)(=O)=O)S(O)(=O)=O)=C/C=C1/N(CCCCS(O)(=O)=O)c2ccc3c(cc(cc3c2C1(C)C)S(O)(=O)=O)S([O-])(=O)=O)P(O)(=O)C[C@@H](CCc1ccccc1)C(O)=O |r,c:54|