Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
cGMP-specific 3',5'-cyclic phosphodiesterase
Ligand
BDBM50111902
Substrate
n/a
Meas. Tech.
ChEMBL_304855 (CHEMBL828414)
IC50
2±n/a nM
Citation
 Haning, H; Niewöhner, U; Schenke, T; Lampe, T; Hillisch, A; Bischoff, E Comparison of different heterocyclic scaffolds as substrate analog PDE5 inhibitors. Bioorg Med Chem Lett 15:3900-7 (2005) [PubMed] Article
Haning, H; Niewöhner, U; Schenke, T; Lampe, T; Hillisch, A; Bischoff, E Comparison of different heterocyclic scaffolds as substrate analog PDE5 inhibitors. Bioorg Med Chem Lett 15:3900-7 (2005) [PubMed] Article More Info.:
Target
Name:
cGMP-specific 3',5'-cyclic phosphodiesterase
Synonyms:
3',5'-cyclic phosphodiesterase | CGB-PDE | PDE5 | PDE5A | PDE5A_HUMAN | Phosphodiesterase 2 and 5 (PDE2 and PDE5) | Phosphodiesterase 5 (PDE5) | Phosphodiesterase 5A | Phosphodiesterase 5A (PDE5A) | cGMP-binding cGMP-specific phosphodiesterase | cGMP-specific 3',5'-cyclic phosphodiesterase
Type:
Protein
Mol. Mass.:
99975.83
Organism:
Homo sapiens (Human)
Description:
O76074
Residue:
875
Sequence:
MERAGPSFGQQRQQQQPQQQKQQQRDQDSVEAWLDDHWDFTFSYFVRKATREMVNAWFAERVHTIPVCKEGIRGHTESCSCPLQQSPRADNSAPGTPTRKISASEFDRPLRPIVVKDSEGTVSFLSDSEKKEQMPLTPPRFDHDEGDQCSRLLELVKDISSHLDVTALCHKIFLHIHGLISADRYSLFLVCEDSSNDKFLISRLFDVAEGSTLEEVSNNCIRLEWNKGIVGHVAALGEPLNIKDAYEDPRFNAEVDQITGYKTQSILCMPIKNHREEVVGVAQAINKKSGNGGTFTEKDEKDFAAYLAFCGIVLHNAQLYETSLLENKRNQVLLDLASLIFEEQQSLEVILKKIAATIISFMQVQKCTIFIVDEDCSDSFSSVFHMECEELEKSSDTLTREHDANKINYMYAQYVKNTMEPLNIPDVSKDKRFPWTTENTGNVNQQCIRSLLCTPIKNGKKNKVIGVCQLVNKMEENTGKVKPFNRNDEQFLEAFVIFCGLGIQNTQMYEAVERAMAKQMVTLEVLSYHASAAEEETRELQSLAAAVVPSAQTLKITDFSFSDFELSDLETALCTIRMFTDLNLVQNFQMKHEVLCRWILSVKKNYRKNVAYHNWRHAFNTAQCMFAALKAGKIQNKLTDLEILALLIAALSHDLDHRGVNNSYIQRSEHPLAQLYCHSIMEHHHFDQCLMILNSPGNQILSGLSIEEYKTTLKIIKQAILATDLALYIKRRGEFFELIRKNQFNLEDPHQKELFLAMLMTACDLSAITKPWPIQQRIAELVATEFFDQGDRERKELNIEPTDLMNREKKNKIPSMQVGFIDAICLQLYEALTHVSEDCFPLLDGCRKNRQKWQALAEQQEKMLINGESGQAKRN
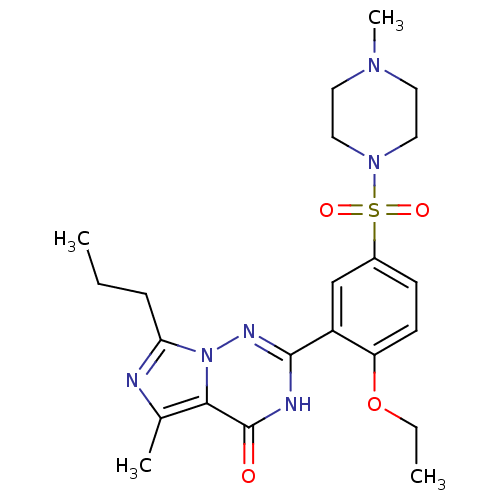
Inhibitor
Name:
BDBM50111902
Synonyms:
2-(2-ethoxy-5-(4-methylpiperazin-1-ylsulfonyl)phenyl)-5-methyl-7-propylimidazo[1,5-f][1,2,4]triazin-4(3H)-one | 2-[2-Ethoxy-5-(4-methyl-piperazine-1-sulfonyl)-phenyl]-5-methyl-7-propyl-3H-imidazo[5,1-f][1,2,4]triazin-4-one | CHEMBL168949
Type:
Small organic molecule
Emp. Form.:
C22H30N6O4S
Mol. Mass.:
474.576
SMILES:
CCCc1nc(C)c2n1nc([nH]c2=O)-c1cc(ccc1OCC)S(=O)(=O)N1CCN(C)CC1