Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Dipeptidyl peptidase 2
Ligand
BDBM50338472
Substrate
n/a
Meas. Tech.
ChEMBL_728290 (CHEMBL1687447)
IC50
58000±n/a nM
Citation
 Chen, P; Caldwell, CG; Ashton, W; Wu, JK; He, H; Lyons, KA; Thornberry, NA; Weber, AE Synthesis and evaluation of [(1R)-1-amino-2-(2,5-difluorophenyl)ethyl]cyclohexanes and 4-[(1R)-1-amino-2-(2,5-difluorophenyl)ethyl]piperidines as DPP-4 inhibitors. Bioorg Med Chem Lett 21:1880-6 (2011) [PubMed] Article
Chen, P; Caldwell, CG; Ashton, W; Wu, JK; He, H; Lyons, KA; Thornberry, NA; Weber, AE Synthesis and evaluation of [(1R)-1-amino-2-(2,5-difluorophenyl)ethyl]cyclohexanes and 4-[(1R)-1-amino-2-(2,5-difluorophenyl)ethyl]piperidines as DPP-4 inhibitors. Bioorg Med Chem Lett 21:1880-6 (2011) [PubMed] Article More Info.:
Target
Name:
Dipeptidyl peptidase 2
Synonyms:
DAP II | DPP2 | DPP2_HUMAN | DPP7 | Dipeptidyl aminopeptidase II | Dipeptidyl peptidase 2 (DPP II) | Dipeptidyl peptidase 2 (DPP2) | Dipeptidyl peptidase II (DDP-II) | Dipeptidyl peptidase II (DPP II) | Dipeptidyl peptidase II (DPP2) | Dipeptidyl peptidase II and dipeptidyl peptidase IV (DPP2 and DPP4) | QPP | carboxytripeptidase | dipeptidyl arylamidase II | dipeptidyl(amino)peptidase II | dipeptidylarylamidase
Type:
Protein
Mol. Mass.:
54339.29
Organism:
Homo sapiens (Human)
Description:
Q9UHL4
Residue:
492
Sequence:
MGSAPWAPVLLLALGLRGLQAGARRAPDPGFQERFFQQRLDHFNFERFGNKTFPQRFLVSDRFWVRGEGPIFFYTGNEGDVWAFANNSAFVAELAAERGALLVFAEHRYYGKSLPFGAQSTQRGHTELLTVEQALADFAELLRALRRDLGAQDAPAIAFGGSYGGMLSAYLRMKYPHLVAGALAASAPVLAVAGLGDSNQFFRDVTADFEGQSPKCTQGVREAFRQIKDLFLQGAYDTVRWEFGTCQPLSDEKDLTQLFMFARNAFTVLAMMDYPYPTDFLGPLPANPVKVGCDRLLSEAQRITGLRALAGLVYNASGSEHCYDIYRLYHSCADPTGCGTGPDARAWDYQACTEINLTFASNNVTDMFPDLPFTDELRQRYCLDTWGVWPRPDWLLTSFWGGDLRAASNIIFSNGNLDPWAGGGIRRNLSASVIAVTIQGGAHHLDLRASHPEDPASVVEARKLEATIIGEWVKAARREQQPALRGGPRLSL
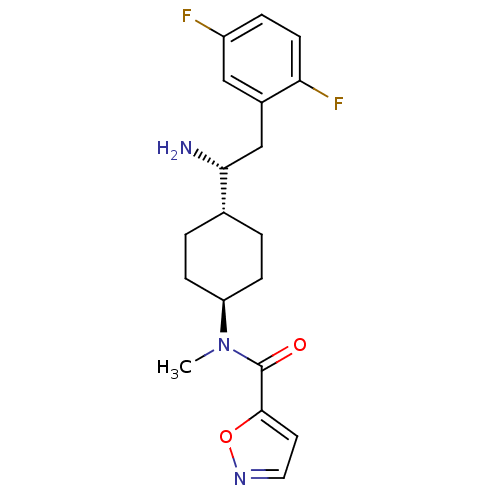
Inhibitor
Name:
BDBM50338472
Synonyms:
CHEMBL1683106 | trans-N-(4-((R)-1-amino-2-(2,5-difluorophenyl)ethyl)cyclohexyl)-N-methylisoxazole-5-carboxamide
Type:
Small organic molecule
Emp. Form.:
C19H23F2N3O2
Mol. Mass.:
363.4016
SMILES:
CN([C@H]1CC[C@@H](CC1)[C@H](N)Cc1cc(F)ccc1F)C(=O)c1ccno1 |r,wU:8.9,2.1,wD:5.5,(34.24,-23.67,;34.24,-22.13,;32.91,-21.35,;31.57,-22.12,;30.25,-21.35,;30.25,-19.82,;31.58,-19.04,;32.91,-19.81,;28.92,-19.05,;28.91,-17.51,;27.58,-19.82,;26.25,-19.05,;26.24,-17.5,;24.91,-16.74,;24.9,-15.2,;23.58,-17.51,;23.58,-19.05,;24.91,-19.82,;24.91,-21.36,;35.58,-21.36,;35.58,-19.82,;36.91,-22.13,;37.07,-23.66,;38.58,-23.98,;39.35,-22.65,;38.32,-21.5,)|