Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Tyrosine-protein kinase ZAP-70
Ligand
BDBM4814
Substrate
n/a
Meas. Tech.
ChEMBL_774224 (CHEMBL1908441)
Kd
>10000±n/a nM
Citation
 Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol 29:1046-51 (2011) [PubMed] Article
Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol 29:1046-51 (2011) [PubMed] Article More Info.:
Target
Name:
Tyrosine-protein kinase ZAP-70
Synonyms:
70 kDa zeta-associated protein | SRK | Syk-related tyrosine kinase | Tyrosine Kinase ZAP-70 | Tyrosine-protein kinase ZAP-70 | Tyrosine-protein kinase ZAP-70 (Syk) | Tyrosine-protein kinase ZAP-70 (ZAP70) | Tyrosine-protein kinase ZAP70 | ZAP70 | ZAP70_HUMAN | Zeta-chain (TCR) associated protein kinase 70kDa
Type:
Enzyme
Mol. Mass.:
69881.61
Organism:
Homo sapiens (Human)
Description:
ZAP-70 SH2 domain was expressed and purified from E. coli using O-phospho-L-tyrosine-agarose column.
Residue:
619
Sequence:
MPDPAAHLPFFYGSISRAEAEEHLKLAGMADGLFLLRQCLRSLGGYVLSLVHDVRFHHFPIERQLNGTYAIAGGKAHCGPAELCEFYSRDPDGLPCNLRKPCNRPSGLEPQPGVFDCLRDAMVRDYVRQTWKLEGEALEQAIISQAPQVEKLIATTAHERMPWYHSSLTREEAERKLYSGAQTDGKFLLRPRKEQGTYALSLIYGKTVYHYLISQDKAGKYCIPEGTKFDTLWQLVEYLKLKADGLIYCLKEACPNSSASNASGAAAPTLPAHPSTLTHPQRRIDTLNSDGYTPEPARITSPDKPRPMPMDTSVYESPYSDPEELKDKKLFLKRDNLLIADIELGCGNFGSVRQGVYRMRKKQIDVAIKVLKQGTEKADTEEMMREAQIMHQLDNPYIVRLIGVCQAEALMLVMEMAGGGPLHKFLVGKREEIPVSNVAELLHQVSMGMKYLEEKNFVHRDLAARNVLLVNRHYAKISDFGLSKALGADDSYYTARSAGKWPLKWYAPECINFRKFSSRSDVWSYGVTMWEALSYGQKPYKKMKGPEVMAFIEQGKRMECPPECPPELYALMSDCWIYKWEDRPDFLTVEQRMRACYYSLASKVEGPPGSTQKAEAACA
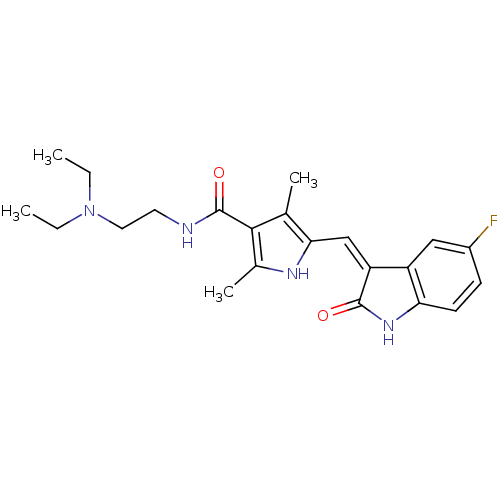
Inhibitor
Name:
BDBM4814
Synonyms:
CHEMBL535 | N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carboxamide | N-[2-(diethylamino)ethyl]-5-{[(3Z)-5-fluoro-2-oxo-2,3-dihydro-1H-indol-3-ylidene]methyl}-2,4-dimethyl-1H-pyrrole-3-carboxamide | SU11248 | SUNITINIB | SUNITINIB MALATE | US10464902, Sunitinib | US20240058459, Compound Sunit-inib | US9163010, Sunitinib | US9914707, SU11248
Type:
Small organic molecule
Emp. Form.:
C22H27FN4O2
Mol. Mass.:
398.4738
SMILES:
CCN(CC)CCNC(=O)c1c(C)[nH]c(\C=C2/C(=O)Nc3ccc(F)cc23)c1C