Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Mitogen-activated protein kinase 10
Ligand
BDBM13216
Substrate
n/a
Meas. Tech.
ChEMBL_586459 (CHEMBL1063713)
Kd
>10000±n/a nM
Citation
 Karaman, MW; Herrgard, S; Treiber, DK; Gallant, P; Atteridge, CE; Campbell, BT; Chan, KW; Ciceri, P; Davis, MI; Edeen, PT; Faraoni, R; Floyd, M; Hunt, JP; Lockhart, DJ; Milanov, ZV; Morrison, MJ; Pallares, G; Patel, HK; Pritchard, S; Wodicka, LM; Zarrinkar, PP A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol 26:127-32 (2008) [PubMed] Article
Karaman, MW; Herrgard, S; Treiber, DK; Gallant, P; Atteridge, CE; Campbell, BT; Chan, KW; Ciceri, P; Davis, MI; Edeen, PT; Faraoni, R; Floyd, M; Hunt, JP; Lockhart, DJ; Milanov, ZV; Morrison, MJ; Pallares, G; Patel, HK; Pritchard, S; Wodicka, LM; Zarrinkar, PP A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol 26:127-32 (2008) [PubMed] Article More Info.:
Target
Name:
Mitogen-activated protein kinase 10
Synonyms:
JNK3 | JNK3A | MAP kinase p49 3F12 | MAPK10 | MK10_HUMAN | Mitogen-Activated Protein Kinase 10 (JNK3) | Mitogen-activated protein kinase 10 (Stress-activated protein kinase JNK3) (c-Jun N-terminal kinase 3) (MAP kinase p49 3F12) | Mitogen-activated protein kinase 10/Receptor-interacting serine/threonine-protein kinase 1 | PRKM10 | SAPK1B | Stress-activated protein kinase JNK3 | c-Jun N-terminal kinase 3 (JNK3)
Type:
Enzyme
Mol. Mass.:
52586.89
Organism:
Homo sapiens (Human)
Description:
n/a
Residue:
464
Sequence:
MSLHFLYYCSEPTLDVKIAFCQGFDKQVDVSYIAKHYNMSKSKVDNQFYSVEVGDSTFTVLKRYQNLKPIGSGAQGIVCAAYDAVLDRNVAIKKLSRPFQNQTHAKRAYRELVLMKCVNHKNIISLLNVFTPQKTLEEFQDVYLVMELMDANLCQVIQMELDHERMSYLLYQMLCGIKHLHSAGIIHRDLKPSNIVVKSDCTLKILDFGLARTAGTSFMMTPYVVTRYYRAPEVILGMGYKENVDIWSVGCIMGEMVRHKILFPGRDYIDQWNKVIEQLGTPCPEFMKKLQPTVRNYVENRPKYAGLTFPKLFPDSLFPADSEHNKLKASQARDLLSKMLVIDPAKRISVDDALQHPYINVWYDPAEVEAPPPQIYDKQLDEREHTIEEWKELIYKEVMNSEEKTKNGVVKGQPSPSGAAVNSSESLPPSSSVNDISSMSTDQTLASDTDSSLEASAGPLGCCR
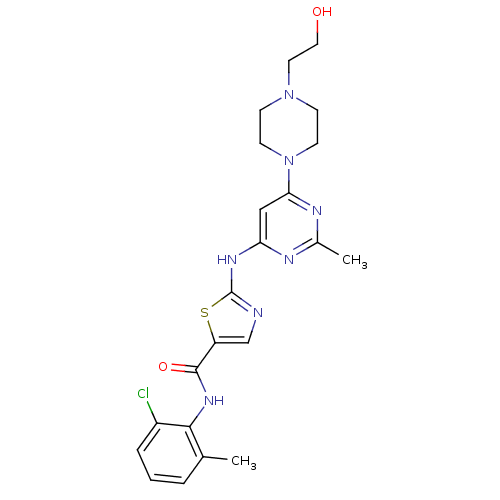
Inhibitor
Name:
BDBM13216
Synonyms:
BMS-354825 | CHEMBL1421 | DASATINIB | N-(2-Chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1-piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide | N-(2-chloro-6-methylphenyl)-2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-1,3-thiazole-5-carboxamide | US10294227, Code Dasatinib | US20230348453, Compound A8 | cid_3062316 | med.21724, Compound Dasatinib
Type:
Small organic molecule
Emp. Form.:
C22H26ClN7O2S
Mol. Mass.:
488.006
SMILES:
Cc1nc(Nc2ncc(s2)C(=O)Nc2c(C)cccc2Cl)cc(n1)N1CCN(CCO)CC1