Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Inhibitor of nuclear factor kappa-B kinase subunit beta
Ligand
BDBM25942
Substrate
n/a
Meas. Tech.
ChEMBL_957967 (CHEMBL2378059)
IC50
7.9±n/a nM
Citation
More Info.:
Target
Name:
Inhibitor of nuclear factor kappa-B kinase subunit beta
Synonyms:
I-kappa-B Kinase 2 (IKK-beta) | I-kappa-B kinase 2 | I-kappa-B-kinase beta | I-kappa-B-kinase beta (IKKB) | IKBKB | IKK-B | IKK-beta | IKK2 | IKK2/IKK1 | IKKB | IKKB_HUMAN | Inhibitor of NF-kappa-B kinase alpha/beta | Inhibitor of nuclear factor kappa B kinase beta subunit | NFKBIKB | Nuclear factor NF-kappa-B inhibitor kinase beta
Type:
Serine/threonine-protein kinase
Mol. Mass.:
86554.39
Organism:
Homo sapiens (Human)
Description:
GST-tagged IKK-2 was expressed in High Five cells and purified.
Residue:
756
Sequence:
MSWSPSLTTQTCGAWEMKERLGTGGFGNVIRWHNQETGEQIAIKQCRQELSPRNRERWCLEIQIMRRLTHPNVVAARDVPEGMQNLAPNDLPLLAMEYCQGGDLRKYLNQFENCCGLREGAILTLLSDIASALRYLHENRIIHRDLKPENIVLQQGEQRLIHKIIDLGYAKELDQGSLCTSFVGTLQYLAPELLEQQKYTVTVDYWSFGTLAFECITGFRPFLPNWQPVQWHSKVRQKSEVDIVVSEDLNGTVKFSSSLPYPNNLNSVLAERLEKWLQLMLMWHPRQRGTDPTYGPNGCFKALDDILNLKLVHILNMVTGTIHTYPVTEDESLQSLKARIQQDTGIPEEDQELLQEAGLALIPDKPATQCISDGKLNEGHTLDMDLVFLFDNSKITYETQISPRPQPESVSCILQEPKRNLAFFQLRKVWGQVWHSIQTLKEDCNRLQQGQRAAMMNLLRNNSCLSKMKNSMASMSQQLKAKLDFFKTSIQIDLEKYSEQTEFGITSDKLLLAWREMEQAVELCGRENEVKLLVERMMALQTDIVDLQRSPMGRKQGGTLDDLEEQARELYRRLREKPRDQRTEGDSQEMVRLLLQAIQSFEKKVRVIYTQLSKTVVCKQKALELLPKVEEVVSLMNEDEKTVVRLQEKRQKELWNLLKIACSKVRGPVSGSPDSMNASRLSQPGQLMSQPSTASNSLPEPAKKSEELVAEAHNLCTLLENAIQDTVREQDQSFTALDWSWLQTEEEEHSCLEQAS
Inhibitor
Name:
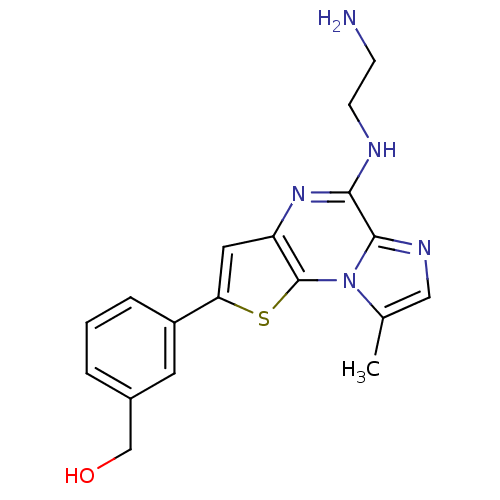
BDBM25942
Synonyms:
(3-{8-[(2-aminoethyl)amino]-12-methyl-3-thia-1,7,10-triazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),4,7,9,11-pentaen-4-yl}phenyl)methanol | imidazothienopyrazine, 14e
Type:
Small organic molecule
Emp. Form.:
C18H19N5OS
Mol. Mass.:
353.441
SMILES:
Cc1cnc2c(NCCN)nc3cc(sc3n12)-c1cccc(CO)c1