Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Potassium voltage-gated channel subfamily D member 3
Ligand
BDBM13216
Substrate
n/a
Meas. Tech.
ChEMBL_1478638 (CHEMBL3430539)
IC50
316228±n/a nM
Citation
 Mirams, GR; Davies, MR; Brough, SJ; Bridgland-Taylor, MH; Cui, Y; Gavaghan, DJ; Abi-Gerges, N Prediction of Thorough QT study results using action potential simulations based on ion channel screens. J Pharmacol Toxicol Methods 70:246-54 (2014) [PubMed] Article
Mirams, GR; Davies, MR; Brough, SJ; Bridgland-Taylor, MH; Cui, Y; Gavaghan, DJ; Abi-Gerges, N Prediction of Thorough QT study results using action potential simulations based on ion channel screens. J Pharmacol Toxicol Methods 70:246-54 (2014) [PubMed] Article More Info.:
Target
Name:
Potassium voltage-gated channel subfamily D member 3
Synonyms:
KCND3 | KCND3_HUMAN | Kv channel-interacting protein 2/Potassium voltage-gated channel subfamily D member 3 | Potassium voltage-gated channel subfamily D member 3 | Voltage-gated potassium channel | Voltage-gated potassium channel subunit Kv4.3
Type:
PROTEIN
Mol. Mass.:
73464.62
Organism:
Homo sapiens (Human)
Description:
ChEMBL_1478652
Residue:
655
Sequence:
MAAGVAAWLPFARAAAIGWMPVANCPMPLAPADKNKRQDELIVLNVSGRRFQTWRTTLERYPDTLLGSTEKEFFFNEDTKEYFFDRDPEVFRCVLNFYRTGKLHYPRYECISAYDDELAFYGILPEIIGDCCYEEYKDRKRENAERLMDDNDSENNQESMPSLSFRQTMWRAFENPHTSTLALVFYYVTGFFIAVSVITNVVETVPCGTVPGSKELPCGERYSVAFFCLDTACVMIFTVEYLLRLFAAPSRYRFIRSVMSIIDVVAIMPYYIGLVMTNNEDVSGAFVTLRVFRVFRIFKFSRHSQGLRILGYTLKSCASELGFLLFSLTMAIIIFATVMFYAEKGSSASKFTSIPASFWYTIVTMTTLGYGDMVPKTIAGKIFGSICSLSGVLVIALPVPVIVSNFSRIYHQNQRADKRRAQKKARLARIRVAKTGSSNAYLHSKRNGLLNEALELTGTPEEEHMGKTTSLIESQHHHLLHCLEKTTGLSYLVDDPLLSVRTSTIKNHEFIDEQMFEQNCMESSMQNYPSTRSPSLSSHPGLTTTCCSRRSKKTTHLPNSNLPATRLRSMQELSTIHIQGSEQPSLTTSRSSLNLKADDGLRPNCKTSQITTAIISIPTPPALTPEGESRPPPASPGPNTNIPSIASNVVKVSAL
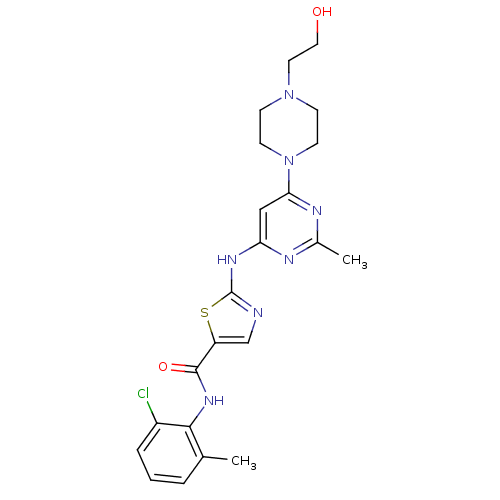
Inhibitor
Name:
BDBM13216
Synonyms:
BMS-354825 | CHEMBL1421 | DASATINIB | N-(2-Chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1-piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide | N-(2-chloro-6-methylphenyl)-2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-1,3-thiazole-5-carboxamide | US10294227, Code Dasatinib | US20230348453, Compound A8 | cid_3062316 | med.21724, Compound Dasatinib
Type:
Small organic molecule
Emp. Form.:
C22H26ClN7O2S
Mol. Mass.:
488.006
SMILES:
Cc1nc(Nc2ncc(s2)C(=O)Nc2c(C)cccc2Cl)cc(n1)N1CCN(CCO)CC1