Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Heat shock cognate 71 kDa protein
Ligand
BDBM50049820
Substrate
n/a
Meas. Tech.
ChEMBL_1588265 (CHEMBL3825930)
Kd
3236±n/a nM
Citation
 Cheeseman, MD; Westwood, IM; Barbeau, O; Rowlands, M; Dobson, S; Jones, AM; Jeganathan, F; Burke, R; Kadi, N; Workman, P; Collins, I; van Montfort, RL; Jones, K Exploiting Protein Conformational Change to Optimize Adenosine-Derived Inhibitors of HSP70. J Med Chem 59:4625-36 (2016) [PubMed] Article
Cheeseman, MD; Westwood, IM; Barbeau, O; Rowlands, M; Dobson, S; Jones, AM; Jeganathan, F; Burke, R; Kadi, N; Workman, P; Collins, I; van Montfort, RL; Jones, K Exploiting Protein Conformational Change to Optimize Adenosine-Derived Inhibitors of HSP70. J Med Chem 59:4625-36 (2016) [PubMed] Article More Info.:
Target
Name:
Heat shock cognate 71 kDa protein
Synonyms:
HSC70 | HSP73 | HSP7C_HUMAN | HSPA10 | HSPA8 | heat shock 70kDa protein 8 isoform 1 | heat shock 70kDa protein 8 isoform 2
Type:
Enzyme Catalytic Domain
Mol. Mass.:
70887.67
Organism:
Homo sapiens (Human)
Description:
gi_5729877
Residue:
646
Sequence:
MSKGPAVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMNPTNTVFDAKRLIGRRFDDAVVQSDMKHWPFMVVNDAGRPKVQVEYKGETKSFYPEEVSSMVLTKMKEIAEAYLGKTVTNAVVTVPAYFNDSQRQATKDAGTIAGLNVLRIINEPTAAAIAYGLDKKVGAERNVLIFDLGGGTFDVSILTIEDGIFEVKSTAGDTHLGGEDFDNRMVNHFIAEFKRKHKKDISENKRAVRRLRTACERAKRTLSSSTQASIEIDSLYEGIDFYTSITRARFEELNADLFRGTLDPVEKALRDAKLDKSQIHDIVLVGGSTRIPKIQKLLQDFFNGKELNKSINPDEAVAYGAAVQAAILSGDKSENVQDLLLLDVTPLSLGIETAGGVMTVLIKRNTTIPTKQTQTFTTYSDNQPGVLIQVYEGERAMTKDNNLLGKFELTGIPPAPRGVPQIEVTFDIDANGILNVSAVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDEKQRDKVSSKNSLESYAFNMKATVEDEKLQGKINDEDKQKILDKCNEIINWLDKNQTAEKEEFEHQQKELEKVCNPIITKLYQSAGGMPGGMPGGFPGGGAPPSGGASSGPTIEEVD
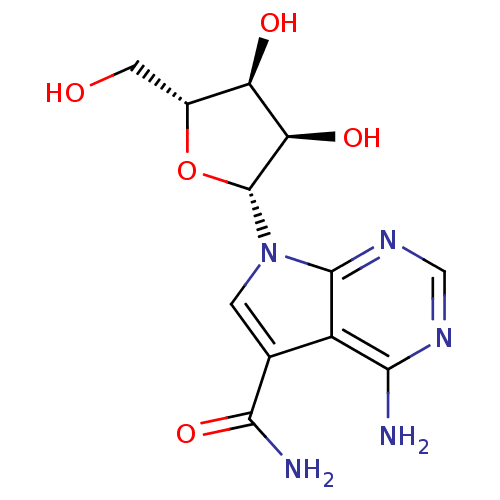
Inhibitor
Name:
BDBM50049820
Synonyms:
4-Amino-5-carboxamide-7-(D-ribofuranosyl)pyrrolo[2,3-d]pyrimidine | 4-Amino-7-((2R,3R,4S,5R)-3,4-dihydroxy-5-hydroxymethyl-tetrahydro-furan-2-yl)-7H-pyrrolo[2,3-d]pyrimidine-5-carboxylic acid amide | 4-Amino-7-(3,4-dihydroxy-5-hydroxymethyl-tetrahydro-furan-2-yl)-7H-pyrrolo[2,3-d]pyrimidine-5-carboxylic acid amide | 4-amino-7-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)-tetrahydrofuran-2-yl)-7H-pyrrolo[2,3-d]pyrimidine-5-carboxamide | CHEMBL101892 | SANGIVAMYCIN | cid_5153
Type:
Small organic molecule
Emp. Form.:
C12H15N5O5
Mol. Mass.:
309.278
SMILES:
NC(=O)c1cn([C@@H]2O[C@H](CO)[C@@H](O)[C@H]2O)c2ncnc(N)c12 |r|