Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Histone-lysine N-methyltransferase EZH2
Ligand
BDBM50193709
Substrate
n/a
Meas. Tech.
ChEMBL_1615977 (CHEMBL3858046)
Ki
0.700000±n/a nM
Citation
 Kung, PP; Rui, E; Bergqvist, S; Bingham, P; Braganza, J; Collins, M; Cui, M; Diehl, W; Dinh, D; Fan, C; Fantin, VR; Gukasyan, HJ; Hu, W; Huang, B; Kephart, S; Krivacic, C; Kumpf, RA; Li, G; Maegley, KA; McAlpine, I; Nguyen, L; Ninkovic, S; Ornelas, M; Ryskin, M; Scales, S; Sutton, S; Tatlock, J; Verhelle, D; Wang, F; Wells, P; Wythes, M; Yamazaki, S; Yip, B; Yu, X; Zehnder, L; Zhang, WG; Rollins, RA; Edwards, M Design and Synthesis of Pyridone-Containing 3,4-Dihydroisoquinoline-1(2H)-ones as a Novel Class of Enhancer of Zeste Homolog 2 (EZH2) Inhibitors. J Med Chem 59:8306-25 (2016) [PubMed] Article
Kung, PP; Rui, E; Bergqvist, S; Bingham, P; Braganza, J; Collins, M; Cui, M; Diehl, W; Dinh, D; Fan, C; Fantin, VR; Gukasyan, HJ; Hu, W; Huang, B; Kephart, S; Krivacic, C; Kumpf, RA; Li, G; Maegley, KA; McAlpine, I; Nguyen, L; Ninkovic, S; Ornelas, M; Ryskin, M; Scales, S; Sutton, S; Tatlock, J; Verhelle, D; Wang, F; Wells, P; Wythes, M; Yamazaki, S; Yip, B; Yu, X; Zehnder, L; Zhang, WG; Rollins, RA; Edwards, M Design and Synthesis of Pyridone-Containing 3,4-Dihydroisoquinoline-1(2H)-ones as a Novel Class of Enhancer of Zeste Homolog 2 (EZH2) Inhibitors. J Med Chem 59:8306-25 (2016) [PubMed] Article More Info.:
Target
Name:
Histone-lysine N-methyltransferase EZH2
Synonyms:
ENX-1 | EZH2 | EZH2_HUMAN | Enhancer of zeste homolog 2 (EZH2) | Histone-lysine N-methyltransferase EZH2 | KMT6 | Lysine N-methyltransferase 6
Type:
Protein
Mol. Mass.:
85367.84
Organism:
Homo sapiens (Human)
Description:
Q15910
Residue:
746
Sequence:
MGQTGKKSEKGPVCWRKRVKSEYMRLRQLKRFRRADEVKSMFSSNRQKILERTEILNQEWKQRRIQPVHILTSVSSLRGTRECSVTSDLDFPTQVIPLKTLNAVASVPIMYSWSPLQQNFMVEDETVLHNIPYMGDEVLDQDGTFIEELIKNYDGKVHGDRECGFINDEIFVELVNALGQYNDDDDDDDGDDPEEREEKQKDLEDHRDDKESRPPRKFPSDKIFEAISSMFPDKGTAEELKEKYKELTEQQLPGALPPECTPNIDGPNAKSVQREQSLHSFHTLFCRRCFKYDCFLHPFHATPNTYKRKNTETALDNKPCGPQCYQHLEGAKEFAAALTAERIKTPPKRPGGRRRGRLPNNSSRPSTPTINVLESKDTDSDREAGTETGGENNDKEEEEKKDETSSSSEANSRCQTPIKMKPNIEPPENVEWSGAEASMFRVLIGTYYDNFCAIARLIGTKTCRQVYEFRVKESSIIAPAPAEDVDTPPRKKKRKHRLWAAHCRKIQLKKDGSSNHVYNYQPCDHPRQPCDSSCPCVIAQNFCEKFCQCSSECQNRFPGCRCKAQCNTKQCPCYLAVRECDPDLCLTCGAADHWDSKNVSCKNCSIQRGSKKHLLLAPSDVAGWGIFIKDPVQKNEFISEYCGEIISQDEADRRGKVYDKYMCSFLFNLNNDFVVDATRKGNKIRFANHSVNPNCYAKVMMVNGDHRIGIFAKRAIQTGEELFFDYRYSQADALKYVGIEREMEIP
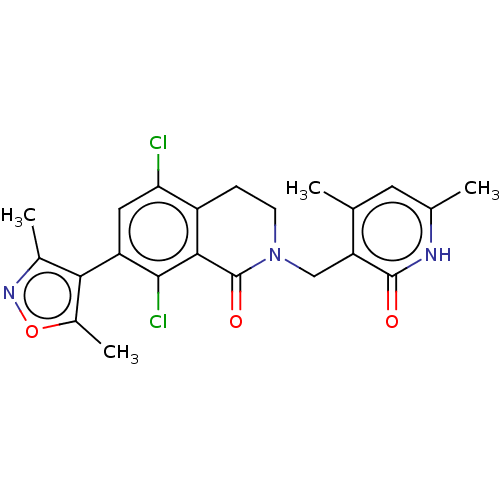
Inhibitor
Name:
BDBM50193709
Synonyms:
CHEMBL3911017
Type:
Small organic molecule
Emp. Form.:
C22H21Cl2N3O3
Mol. Mass.:
446.326
SMILES:
Cc1noc(C)c1-c1cc(Cl)c2CCN(Cc3c(C)cc(C)[nH]c3=O)C(=O)c2c1Cl |(59.06,-26.01,;58.74,-27.52,;59.77,-28.66,;59,-30,;57.5,-29.68,;56.35,-30.71,;57.33,-28.15,;55.99,-27.38,;54.66,-28.14,;53.34,-27.37,;52.01,-28.14,;53.35,-25.85,;52.02,-25.09,;52.02,-23.55,;53.35,-22.77,;53.35,-21.23,;52.02,-20.46,;52.02,-18.92,;53.36,-18.16,;50.7,-18.15,;49.36,-18.92,;48.03,-18.14,;49.35,-20.46,;50.69,-21.24,;50.69,-22.78,;54.68,-23.55,;56.01,-22.79,;54.68,-25.09,;55.99,-25.86,;57.33,-25.09,)|