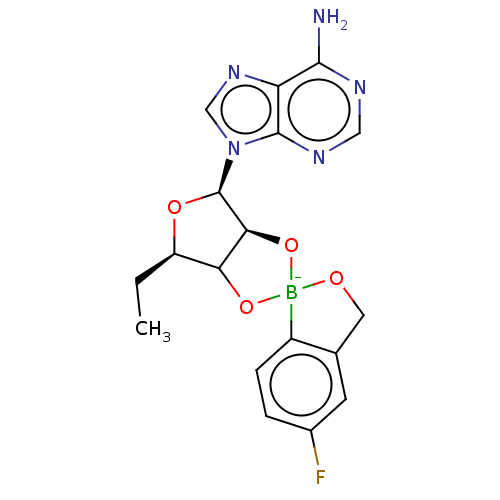
BDBM163674 AN2690-AMP
SMILES CC[C@H]1O[C@H]([C@H]2O[B-]3(OCc4cc(F)ccc34)OC12)n1cnc2c(N)ncnc12
InChI Key InChIKey=WTUJYZMKPXKECI-TZVUDSRHSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 163674
Found 3 hits for monomerid = 163674
Affinity DataKd: 700nMpH: 8.0 T: 2°CAssay Description:ITC was performed using LeuRS CP1 domain wild-type, K510E mutant, or K510A mutant against AN2690-AMP at 25 °C by an ITC200 system (MicroCal, GE Healt...More data for this Ligand-Target Pair
Affinity DataKd: 4.60E+4nMpH: 8.0 T: 2°CAssay Description:ITC was performed using LeuRS CP1 domain wild-type, K510E mutant, or K510A mutant against AN2690-AMP at 25 °C by an ITC200 system (MicroCal, GE Healt...More data for this Ligand-Target Pair
Affinity DataKd: 2.10E+4nMpH: 8.0 T: 2°CAssay Description:ITC was performed using LeuRS CP1 domain wild-type, K510E mutant, or K510A mutant against AN2690-AMP at 25 °C by an ITC200 system (MicroCal, GE Healt...More data for this Ligand-Target Pair
Activity Spreadsheet -- ITC Data from BindingDB
 Found 3 hits for monomerid = 163674
Found 3 hits for monomerid = 163674
ITC DataΔG°: -8.36kcal/mole −TΔS°: 7.12kcal/mole ΔH°: -1.00kcal/mole logk: 1.36E+6
pH: 8.0 T: 25.00°C
pH: 8.0 T: 25.00°C
ITC DataΔG°: -6.45kcal/mole −TΔS°: 4.77kcal/mole ΔH°: -1.50kcal/mole logk: 5.38E+4
pH: 8.0 T: 25.00°C
pH: 8.0 T: 25.00°C
ITC DataΔG°: -5.97kcal/mole −TΔS°: 4.27kcal/mole ΔH°: -1.50kcal/mole logk: 2.40E+4
pH: 8.0 T: 25.00°C
pH: 8.0 T: 25.00°C