

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM36122 Z-L-Glu-L-Tyr
SMILES: OC(=O)CC[C@H](NC(=O)OCc1ccccc1)C(=O)N[C@@H](Cc1ccc(O)cc1)C(O)=O
InChI Key: InChIKey=XLUMOZQZGPJGTL-UHFFFAOYSA-N
Data: 6 ITC
| Cell (A) | Syringe (B) | Cell Links | Syringe Links | Cell + Syr Links | ΔG° kcal/mole | -TΔS° kcal/mole | ΔH° kcal/mole | log K | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|
BDBM4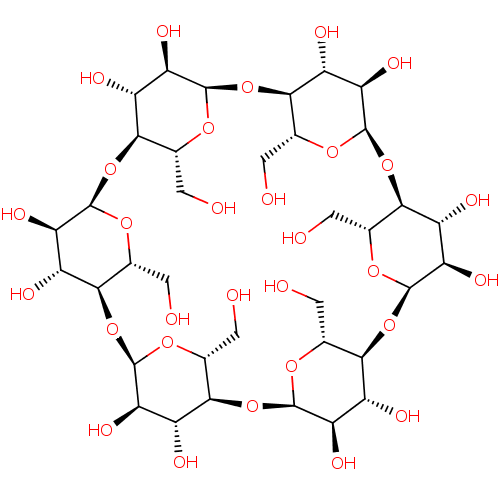 | BDBM36122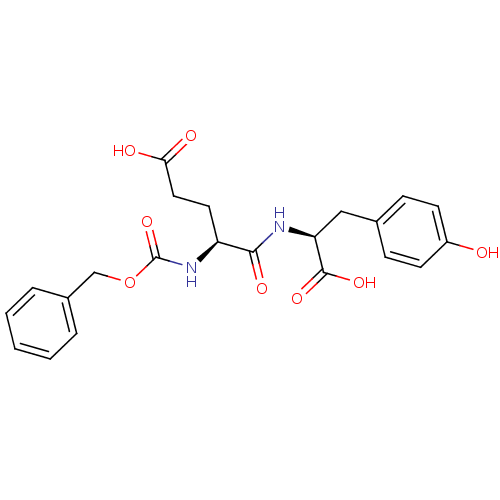 | -1.70 | 2.70 | -4.39 | 1.24 | 6.90 | 25 | |||
TBA | Details of this binding reaction | |||||||||
BDBM11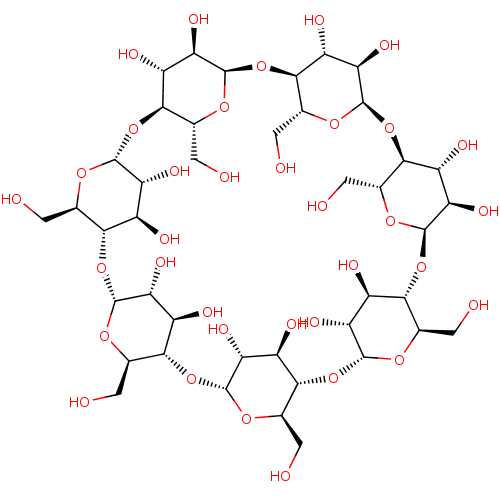 | BDBM36122 | -2.71 | 0.477 | -3.18 | 1.99 | 6.90 | 25 | |||
TBA | Details of this binding reaction | |||||||||
BDBM36123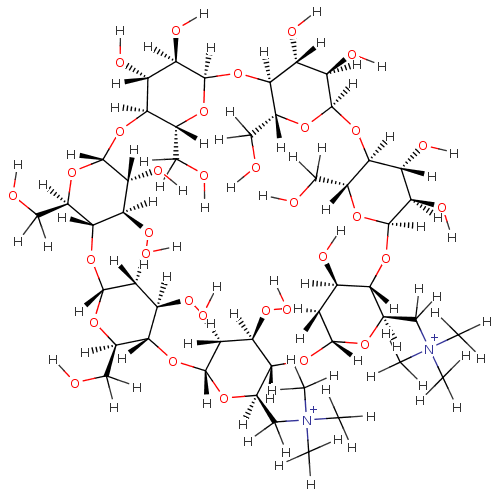 | BDBM36122 | -2.78 | 0.691 | -3.46 | 2.04 | 6.90 | 25 | |||
TBA | Details of this binding reaction | |||||||||
BDBM36124 | BDBM36122 | -2.85 | 0.385 | -3.22 | 2.09 | 6.90 | 25 | |||
TBA | Details of this binding reaction | |||||||||
BDBM36125 | BDBM36122 | -2.78 | 0.456 | -3.22 | 2.04 | 6.90 | 25 | |||
TBA | Details of this binding reaction | |||||||||
BDBM36126 | BDBM36122 | -2.18 | 3.15 | -5.33 | 1.60 | 6.90 | 25 | |||
TBA | Details of this binding reaction | |||||||||