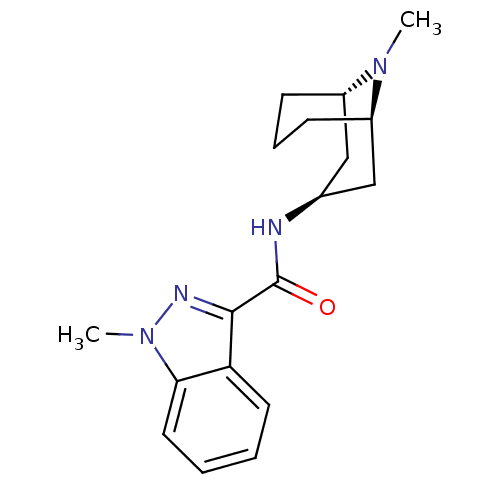
BDBM50449636 BRL-43694::GRANISETRON::Kytril::LY-278584::Sancuso
SMILES [H][C@]12CCC[C@]([H])(C[C@H](C1)NC(=O)c1nn(C)c3ccccc13)N2C
InChI Key InChIKey=MFWNKCLOYSRHCJ-ZSOGYDGISA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 7 hits for monomerid = 50449636
Found 7 hits for monomerid = 50449636
Affinity DataKi: 0.350nMAssay Description:Ability to displace [3H]granisetron specifically bound to 5-hydroxytryptamine 3 receptor in rat cortical membraneMore data for this Ligand-Target Pair
Affinity DataKi: 0.350nMAssay Description:In vitro affinity for 5-hydroxytryptamine 3 (5-HT3) receptor by displacement of [3H]BRL-43694 from rat entorhinal cortexMore data for this Ligand-Target Pair
Affinity DataKi: 0.5nMAssay Description:Binding affinity for 5-hydroxytryptamine 3 receptor was determined by measuring displacement of [3H]GR-65630 from rat brain corticesMore data for this Ligand-Target Pair
Affinity DataKi: 0.600nMAssay Description:Compound was evaluated for its binding affinity for 5-hydroxytryptamine 3 receptor by measuring displacement [3H]GR-65630 in rat cerebral cortexMore data for this Ligand-Target Pair
Affinity DataKi: 0.790nMAssay Description:Binding affinity against 5-hydroxytryptamine 3 (5-HT3) receptor in rat brain cortical membranes using radioligand [3H]quipazineChecked by AuthorMore data for this Ligand-Target Pair
Affinity DataKi: 2.10nMAssay Description:Binding affinity against radioligand [3H]quipazine labeled 5-hydroxytryptamine 3 receptor sites in neuroblastoma-glioma (NG108-15) cells.More data for this Ligand-Target Pair
TargetPotassium voltage-gated channel subfamily H member 2(Homo sapiens (Human))
Johnson & Johnson Pharmaceutical Research & Development
Curated by ChEMBL
Johnson & Johnson Pharmaceutical Research & Development
Curated by ChEMBL
Affinity DataIC50: 3.80E+3nMAssay Description:Inhibition of partially open human voltage-gated potassium channel subunit Kv11.1 (ERG K+ channel)More data for this Ligand-Target Pair