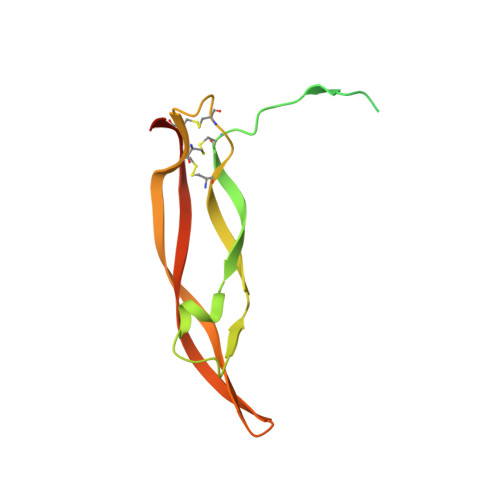
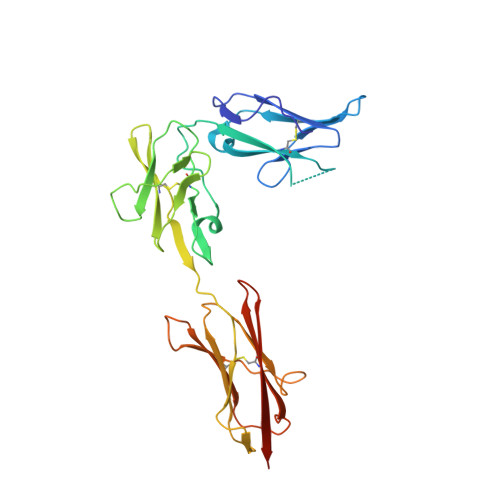
Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex.
Shim, A.H., Liu, H., Focia, P.J., Chen, X., Lin, P.C., He, X.(2010) Proc Natl Acad Sci U S A 107: 11307-11312
- PubMed: 20534510
- DOI: https://doi.org/10.1073/pnas.1000806107
- Primary Citation of Related Structures:
3MJG, 3MJK - PubMed Abstract:
Platelet-derived growth factors (PDGFs) and their receptors (PDGFRs) are prototypic growth factors and receptor tyrosine kinases which have critical functions in development. We show that PDGFs share a conserved region in their prodomain sequences which can remain noncovalently associated with the mature cystine-knot growth factor domain after processing. The structure of the PDGF-A/propeptide complex reveals this conserved, hydrophobic association mode. We also present the structure of the complex between PDGF-B and the first three Ig domains of PDGFRbeta, showing that two PDGF-B protomers clamp PDGFRbeta at their dimerization seam. The PDGF-B:PDGFRbeta interface is predominantly hydrophobic, and PDGFRs and the PDGF propeptides occupy overlapping positions on mature PDGFs, rationalizing the need of propeptides by PDGFs to cover functionally important hydrophobic surfaces during secretion. A large-scale structural organization and rearrangement is observed for PDGF-B upon receptor binding, in which the PDGF-B L1 loop, disordered in the structure of the free form, adopts a highly specific conformation to form hydrophobic interactions with the third Ig domain of PDGFRbeta. Calorimetric data also shows that the membrane-proximal homotypic PDGFRalpha interaction, albeit required for activation, contributes negatively to ligand binding. The structural and biochemical data together offer insights into PDGF-PDGFR signaling, as well as strategies for PDGF-antagonism.
Organizational Affiliation:
Department of Molecular Pharmacology and Biological Chemistry, Northwestern University Feinberg School of Medicine, Searle 8-417, 303 East Chicago Avenue, Chicago, IL 60611, USA.