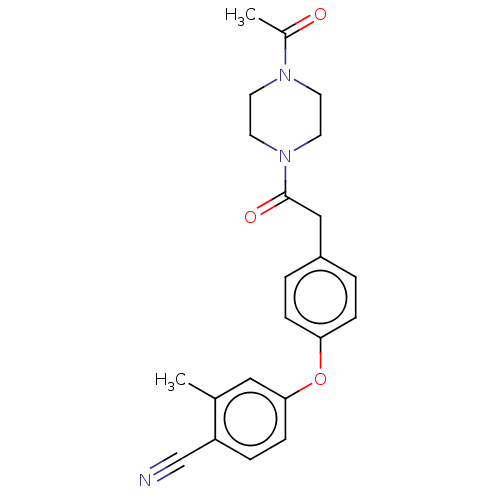
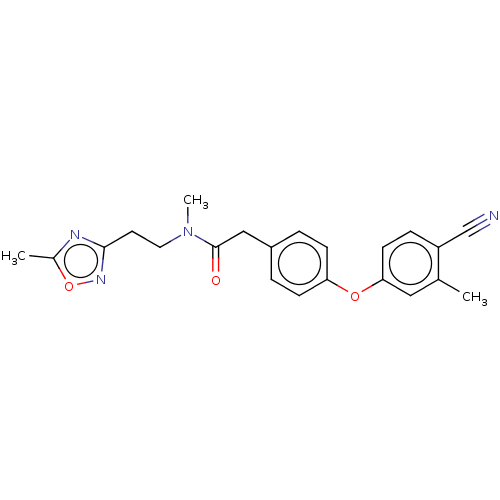
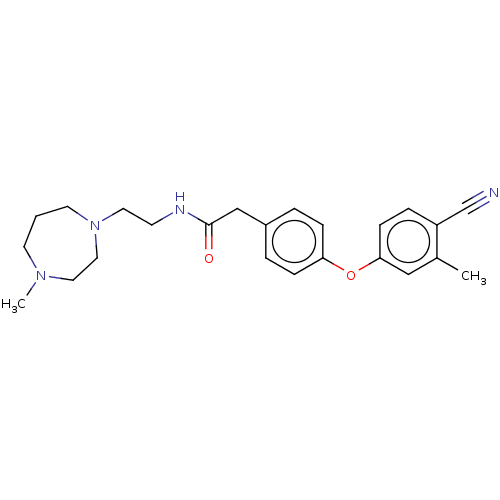
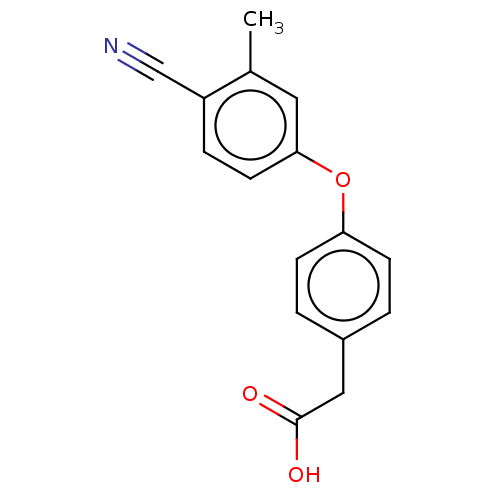
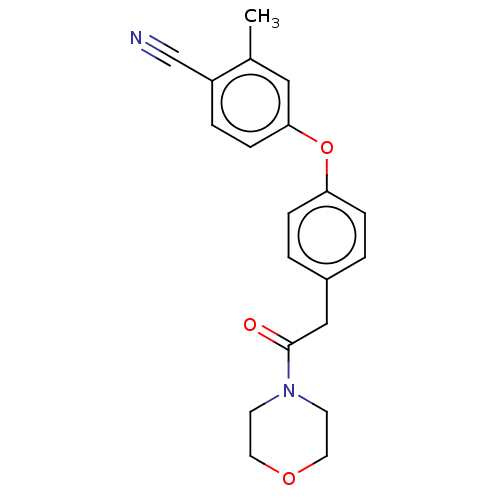
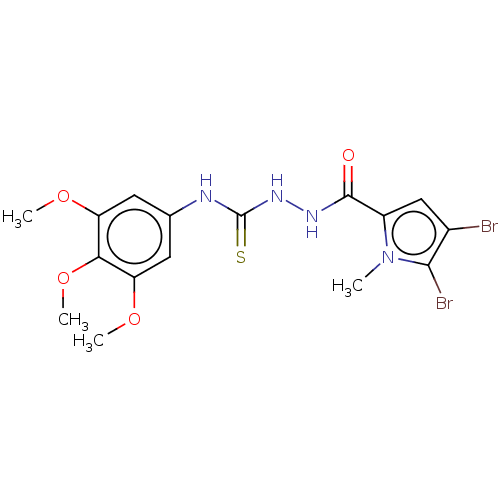
Affinity DataIC50: 1.80E+3nMpH: 8.5 T: 2°CAssay Description:The synthesized compounds were assayed for the inhibitory activities against MMP-2 in 96-well microtiter plates using succinylated gelatin as the sub...More data for this Ligand-Target Pair
Affinity DataIC50: 8.00E+3nMpH: 8.5 T: 2°CAssay Description:The synthesized compounds were assayed for the inhibitory activities against MMP-2 in 96-well microtiter plates using succinylated gelatin as the sub...More data for this Ligand-Target Pair
Affinity DataIC50: 1.37E+4nMpH: 8.5 T: 2°CAssay Description:The synthesized compounds were assayed for the inhibitory activities against MMP-2 in 96-well microtiter plates using succinylated gelatin as the sub...More data for this Ligand-Target Pair
Affinity DataIC50: 1.92E+4nMpH: 8.5 T: 2°CAssay Description:The synthesized compounds were assayed for the inhibitory activities against MMP-2 in 96-well microtiter plates using succinylated gelatin as the sub...More data for this Ligand-Target Pair
Affinity DataIC50: 2.38E+4nMpH: 8.0 T: 2°CAssay Description:The synthesized compounds were assayed for the inhibitory activities against MMP-12 in 96-well microtiter plates using thiopeptolide as the substrate...More data for this Ligand-Target Pair
Affinity DataIC50: 2.90E+4nMpH: 8.0 T: 2°CAssay Description:The synthesized compounds were assayed for the inhibitory activities against MMP-12 in 96-well microtiter plates using thiopeptolide as the substrate...More data for this Ligand-Target Pair
Affinity DataIC50: 4.21E+4nMpH: 8.0 T: 2°CAssay Description:The synthesized compounds were assayed for the inhibitory activities against MMP-12 in 96-well microtiter plates using thiopeptolide as the substrate...More data for this Ligand-Target Pair
Affinity DataIC50: 4.31E+4nMpH: 8.0 T: 2°CAssay Description:The synthesized compounds were assayed for the inhibitory activities against MMP-12 in 96-well microtiter plates using thiopeptolide as the substrate...More data for this Ligand-Target Pair
Affinity DataIC50: 6.03E+4nMpH: 8.5 T: 2°CAssay Description:The synthesized compounds were assayed for the inhibitory activities against MMP-2 in 96-well microtiter plates using succinylated gelatin as the sub...More data for this Ligand-Target Pair
Affinity DataIC50: 8.42E+4nMpH: 8.0 T: 2°CAssay Description:The synthesized compounds were assayed for the inhibitory activities against MMP-12 in 96-well microtiter plates using thiopeptolide as the substrate...More data for this Ligand-Target Pair
TargetThiol:disulfide interchange protein DsbA(Escherichia coli (strain K12))
Monash University (Parkville Campus)
Curated by ChEMBL
Monash University (Parkville Campus)
Curated by ChEMBL
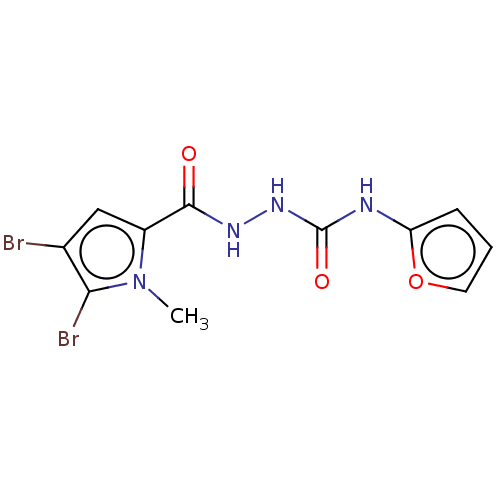
Affinity DataKd: 4.80E+5nMAssay Description:Binding affinity to uniformly labeled 15N oxidized Escherichia coli DsbA by 1H-15N HSQC NMR spectroscopyMore data for this Ligand-Target Pair
TargetThiol:disulfide interchange protein DsbA(Escherichia coli (strain K12))
Monash University (Parkville Campus)
Curated by ChEMBL
Monash University (Parkville Campus)
Curated by ChEMBL
Affinity DataKd: 2.90E+5nMAssay Description:Binding affinity to uniformly labeled 15N oxidized Escherichia coli DsbA by 1H-15N HSQC NMR spectroscopyMore data for this Ligand-Target Pair
TargetThiol:disulfide interchange protein DsbA(Escherichia coli (strain K12))
Monash University (Parkville Campus)
Curated by ChEMBL
Monash University (Parkville Campus)
Curated by ChEMBL
Affinity DataKd: 6.20E+4nMAssay Description:Binding affinity to uniformly labeled 15N oxidized Escherichia coli DsbA by 1H-15N HSQC NMR spectroscopyMore data for this Ligand-Target Pair
TargetThiol:disulfide interchange protein DsbA(Escherichia coli (strain K12))
Monash University (Parkville Campus)
Curated by ChEMBL
Monash University (Parkville Campus)
Curated by ChEMBL
Affinity DataKd: 4.30E+5nMAssay Description:Binding affinity to uniformly labeled 15N oxidized Escherichia coli DsbA by 1H-15N HSQC NMR spectroscopyMore data for this Ligand-Target Pair
TargetThiol:disulfide interchange protein DsbA(Escherichia coli (strain K12))
Monash University (Parkville Campus)
Curated by ChEMBL
Monash University (Parkville Campus)
Curated by ChEMBL
Affinity DataKd: 2.30E+5nMAssay Description:Binding affinity to uniformly labeled 15N oxidized Escherichia coli DsbA by 1H-15N HSQC NMR spectroscopyMore data for this Ligand-Target Pair
TargetThiol:disulfide interchange protein DsbA(Escherichia coli (strain K12))
Monash University (Parkville Campus)
Curated by ChEMBL
Monash University (Parkville Campus)
Curated by ChEMBL
Affinity DataKd: 1.10E+6nMAssay Description:Binding affinity to uniformly labeled 15N oxidized Escherichia coli DsbA by 1H-15N HSQC NMR spectroscopyMore data for this Ligand-Target Pair
TargetThiol:disulfide interchange protein DsbA(Escherichia coli (strain K12))
Monash University (Parkville Campus)
Curated by ChEMBL
Monash University (Parkville Campus)
Curated by ChEMBL
Affinity DataKd: 4.90E+5nMAssay Description:Binding affinity to uniformly labeled 15N oxidized Escherichia coli DsbA by 1H-15N HSQC NMR spectroscopyMore data for this Ligand-Target Pair
TargetThiol:disulfide interchange protein DsbA(Escherichia coli (strain K12))
Monash University (Parkville Campus)
Curated by ChEMBL
Monash University (Parkville Campus)
Curated by ChEMBL
Affinity DataKd: >1.00E+6nMAssay Description:Binding affinity to uniformly labeled 15N oxidized Escherichia coli DsbA by 1H-15N HSQC NMR spectroscopyMore data for this Ligand-Target Pair
TargetThiol:disulfide interchange protein DsbA(Escherichia coli (strain K12))
Monash University (Parkville Campus)
Curated by ChEMBL
Monash University (Parkville Campus)
Curated by ChEMBL
Affinity DataKd: 3.65E+5nMAssay Description:Binding affinity to uniformly labeled 15N oxidized Escherichia coli DsbA by 1H-15N HSQC NMR spectroscopyMore data for this Ligand-Target Pair

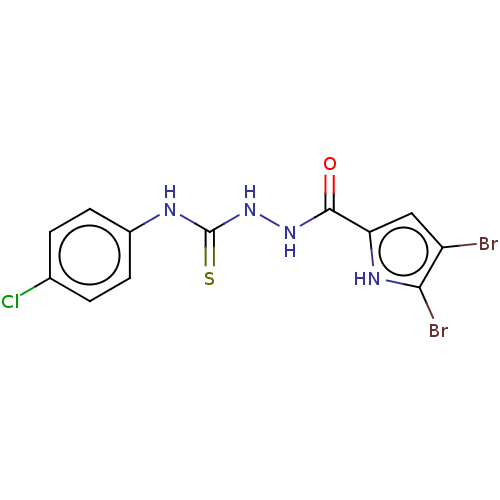
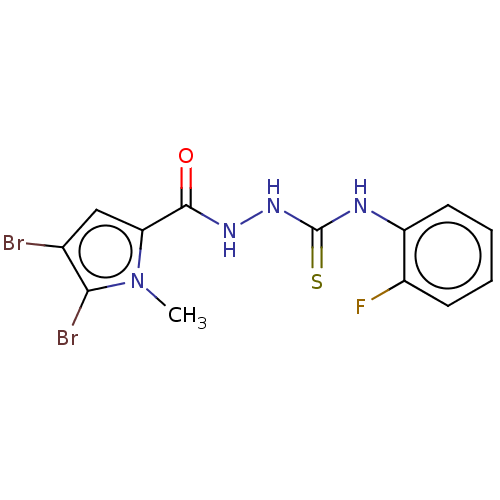
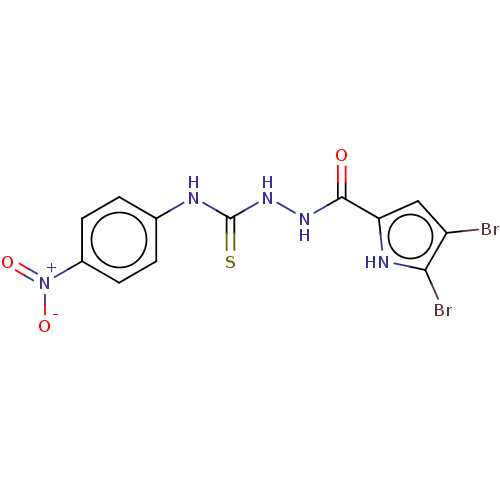

 3D Structure (crystal)
3D Structure (crystal)