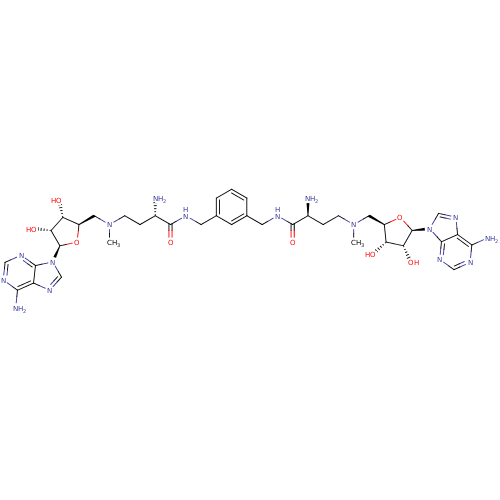
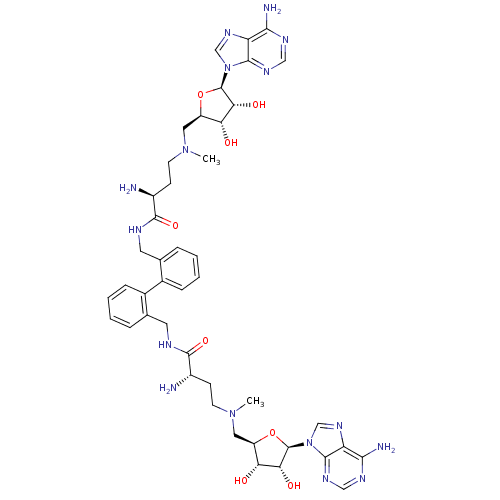
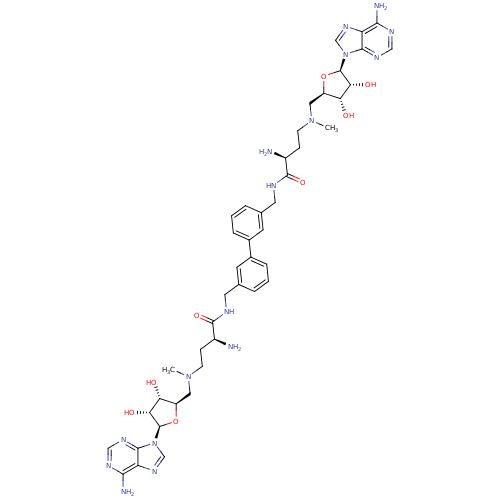
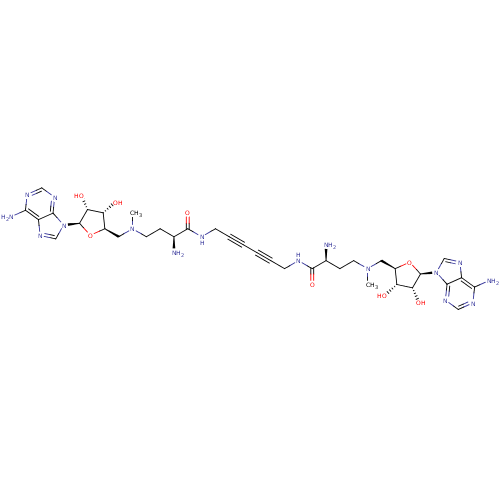
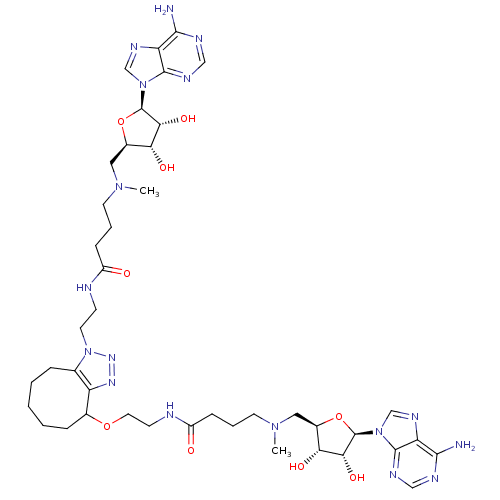
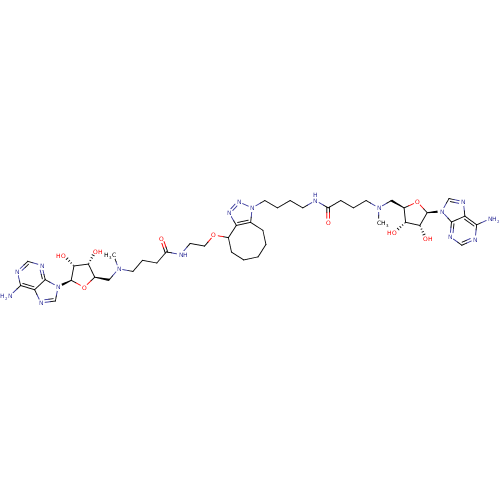
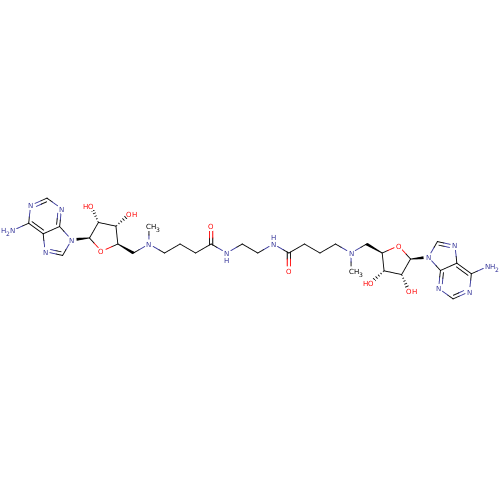
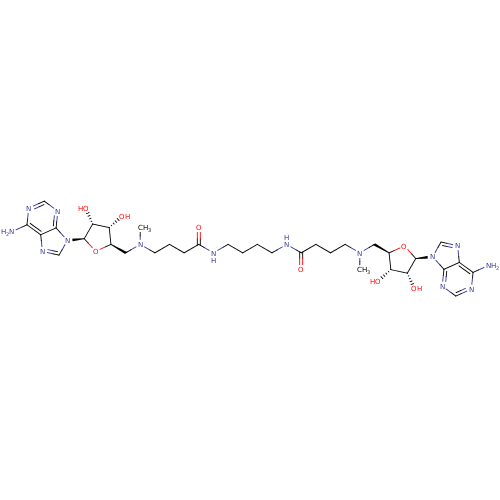
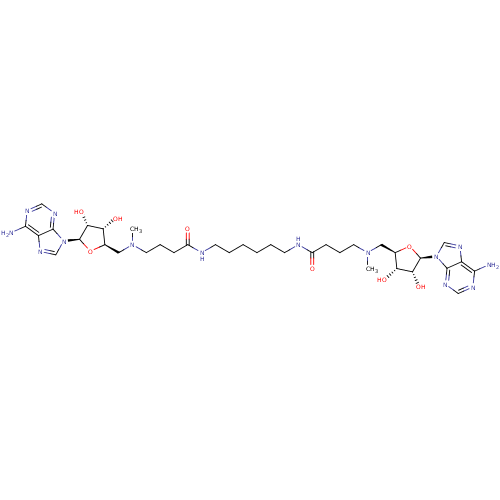
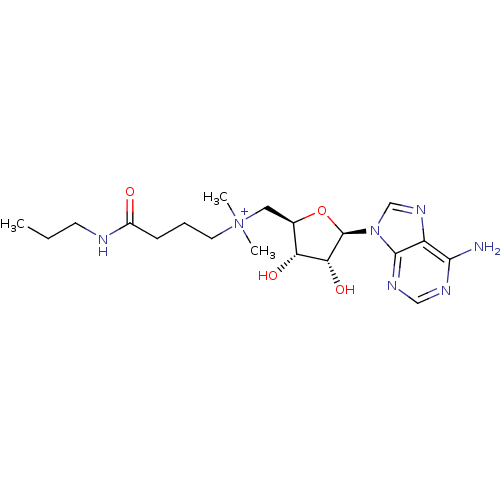
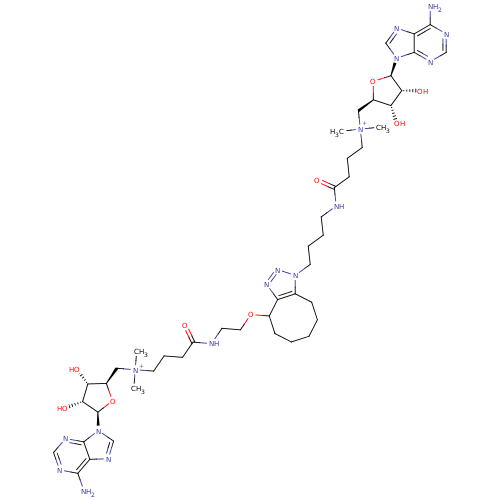
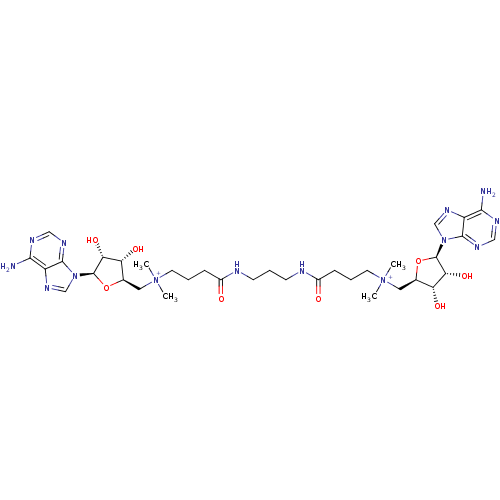
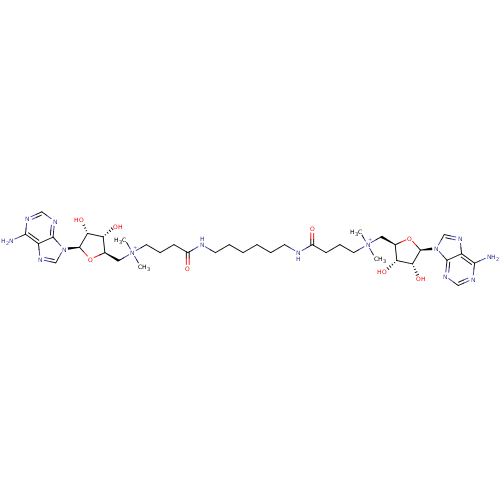
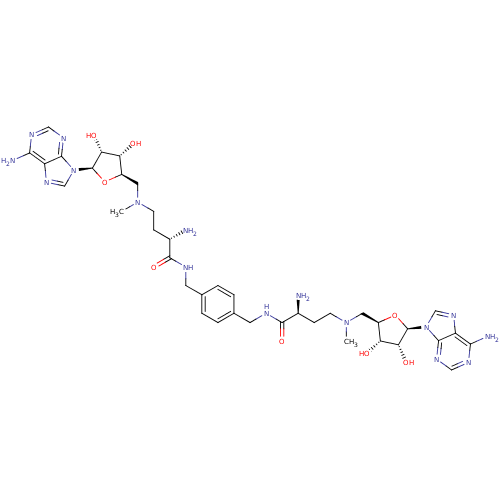
Affinity DataEC50: 120nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataKd: 4nMAssay Description:Binding affinity to Escherichia coli metJ in presence of operator DNA complex by filter binding studyMore data for this Ligand-Target Pair
Affinity DataEC50: 37nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 39nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 75nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 111nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 36nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 15nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 10nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 1.00E+3nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 110nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 76nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 700nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 320nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 220nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 47nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 150nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 50nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 42nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 108nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 51nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 17nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair
Affinity DataEC50: 61nMAssay Description:Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropyMore data for this Ligand-Target Pair

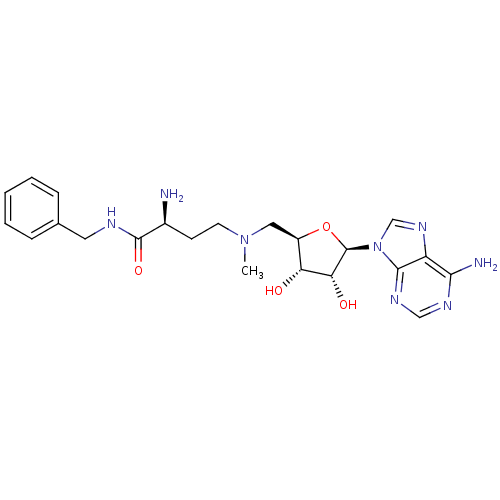
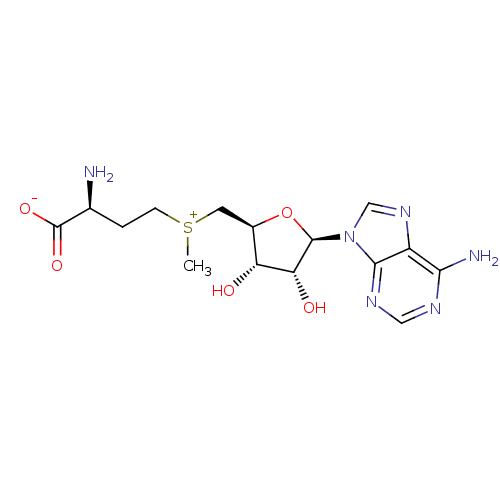
 3D Structure (crystal)
3D Structure (crystal)