Target5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase(Escherichia coli)
Victoria University Of Wellington
Curated by ChEMBL
Victoria University Of Wellington
Curated by ChEMBL
Affinity DataKi: 0.00200nMAssay Description:Inhibition of recombinant Escherichia coli MTAN expressed in Escherichia coli BL-21 DE3 using methylthioadenosine as substrate assessed as inhibition...More data for this Ligand-Target Pair
Target5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase(Escherichia coli (strain K12))
Industrial Research
Industrial Research
Affinity DataKi: 0.0480nM ΔG°: -13.9kcal/molepH: 7.0 T: 2°CAssay Description:Enzyme activity was monitored by absorbance change in the xanthine oxidase coupled assay, which measures the formation of 2,8-dihydroxyadenine at 293...More data for this Ligand-Target Pair
Target5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase(Escherichia coli)
Victoria University Of Wellington
Curated by ChEMBL
Victoria University Of Wellington
Curated by ChEMBL
Affinity DataKi: 0.0480nMAssay Description:Inhibition of recombinant Escherichia coli MTAN expressed in Escherichia coli BL-21 DE3 using methylthioadenosine as substrate by xanthine oxidase co...More data for this Ligand-Target Pair
Target5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase(Escherichia coli (strain K12))
Industrial Research
Industrial Research
Affinity DataKi: 0.0730nM ΔG°: -13.8kcal/molepH: 7.5 T: 2°CAssay Description:Purified MTAN activity in MTAN enzyme inhibition assayMore data for this Ligand-Target Pair
TargetS-methyl-5'-thioadenosine phosphorylase(Homo sapiens (Human))
Victoria University Of Wellington
Curated by ChEMBL
Victoria University Of Wellington
Curated by ChEMBL
Affinity DataKi: 0.0780nMAssay Description:Inhibition of human MTAP using methylthioadenosine as substrate assessed as inhibition constant for slow onset inhibition of enzyme-inhibitor complex...More data for this Ligand-Target Pair
TargetS-methyl-5'-thioadenosine phosphorylase(Homo sapiens (Human))
Victoria University Of Wellington
Curated by ChEMBL
Victoria University Of Wellington
Curated by ChEMBL
Affinity DataKi: 0.0900nMAssay Description:Inhibition of human MTAP assessed as reduction in methylthioadenosine phosphorolysis/hydrolysisMore data for this Ligand-Target Pair
TargetS-methyl-5'-thioadenosine phosphorylase(Homo sapiens (Human))
Victoria University Of Wellington
Curated by ChEMBL
Victoria University Of Wellington
Curated by ChEMBL
Affinity DataKi: 0.530nMAssay Description:Inhibition of human MTAP using methylthioadenosine as substrate by xanthine oxidase coupling enzyme assayMore data for this Ligand-Target Pair
TargetS-methyl-5'-thioadenosine phosphorylase(Homo sapiens (Human))
Victoria University Of Wellington
Curated by ChEMBL
Victoria University Of Wellington
Curated by ChEMBL
Affinity DataKi: 1.70nM ΔG°: -11.8kcal/molepH: 7.0 T: 2°CAssay Description:Enzyme activity was monitored by absorbance change in the xanthine oxidase coupled assay, which measures the formation of 2,8-dihydroxyadenine at 293...More data for this Ligand-Target Pair
Affinity DataKi: 24nM ΔG°: -10.3kcal/molepH: 7.0 T: 2°CAssay Description:Enzyme activity was monitored by absorbance change in the xanthine oxidase coupled assay, which measures the formation of 2,8-dihydroxyadenine at 293...More data for this Ligand-Target Pair

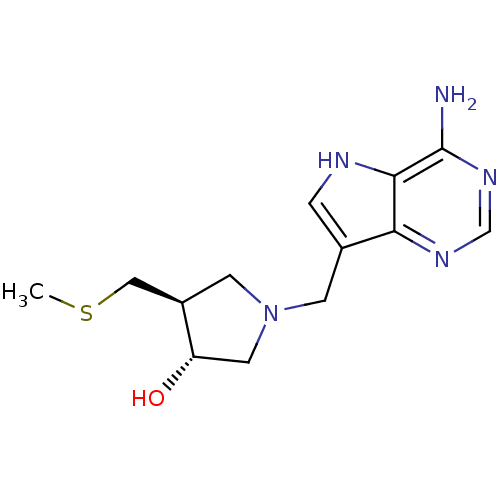
 3D Structure (crystal)
3D Structure (crystal)