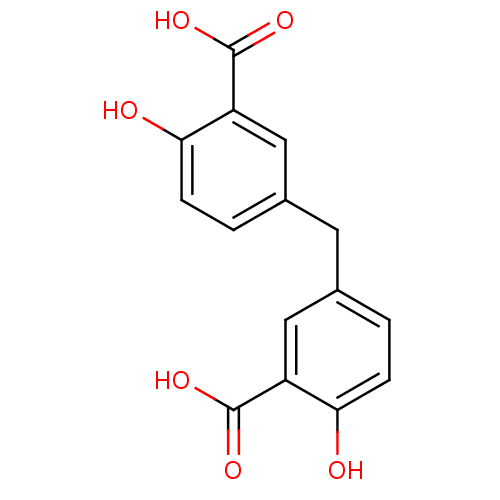
Affinity DataIC50: 8.00E+3nMpH: 7.4 T: 2°CAssay Description:A fluorescence anisotropy (FA)-based biochemical assay that monitors MgrA-DNA binding. To determine the binding affinity of MgrA to the labeled DNA,...More data for this Ligand-Target Pair
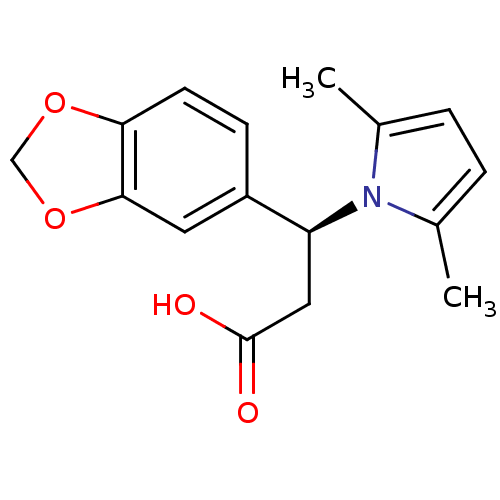
Affinity DataIC50: 2.80E+4nMpH: 7.4 T: 2°CAssay Description:A fluorescence anisotropy (FA)-based biochemical assay that monitors MgrA-DNA binding. To determine the binding affinity of MgrA to the labeled DNA,...More data for this Ligand-Target Pair
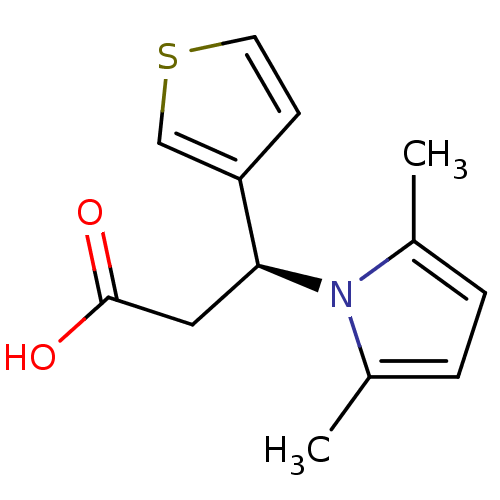
Affinity DataIC50: 3.00E+4nMpH: 7.4 T: 2°CAssay Description:A fluorescence anisotropy (FA)-based biochemical assay that monitors MgrA-DNA binding. To determine the binding affinity of MgrA to the labeled DNA,...More data for this Ligand-Target Pair
Affinity DataIC50: 3.20E+4nMpH: 7.4 T: 2°CAssay Description:A fluorescence anisotropy (FA)-based biochemical assay that monitors MgrA-DNA binding. To determine the binding affinity of MgrA to the labeled DNA,...More data for this Ligand-Target Pair