Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Vasopressin V2 receptor
Ligand
BDBM35706
Substrate
BDBM35667
Meas. Tech.
Binding Affinity Assay (Ki) and cAMP Accumulation Activity Assay (EC50)
pH
7.4±n/a
Temperature
296.15±n/a K
Ki
11±n/a nM
EC50
646±n/a nM
Comments
AMP accumulation max activity = 62.8 %.
Citation
 Tsukamoto, I; Koshio, H; Akamatsu, S; Kuramochi, T; Saitoh, C; Yatsu, T; Yanai-Inamura, H; Kitada, C; Yamamoto, E; Sakamoto, S; Tsukamoto, S Preparation of (4,4-difluoro-1,2,3,4-tetrahydro-5H-1-benzazepin-5-ylidene)acetamide derivatives as novel arginine vasopressin V(2) receptor agonists. Bioorg Med Chem 16:9524-35 (2008) [PubMed] Article
Tsukamoto, I; Koshio, H; Akamatsu, S; Kuramochi, T; Saitoh, C; Yatsu, T; Yanai-Inamura, H; Kitada, C; Yamamoto, E; Sakamoto, S; Tsukamoto, S Preparation of (4,4-difluoro-1,2,3,4-tetrahydro-5H-1-benzazepin-5-ylidene)acetamide derivatives as novel arginine vasopressin V(2) receptor agonists. Bioorg Med Chem 16:9524-35 (2008) [PubMed] Article More Info.:
Target
Name:
Vasopressin V2 receptor
Synonyms:
ADHR | AVPR V2 | AVPR2 | Antidiuretic hormone receptor | DIR | DIR3 | Renal-type arginine vasopressin receptor | V2R | V2R_HUMAN | VASOPRESSIN V2 | Vasopressin V2 receptor | Vasopressin receptor
Type:
Receptor
Mol. Mass.:
40295.28
Organism:
Homo sapiens (Human)
Description:
P30518
Residue:
371
Sequence:
MLMASTTSAVPGHPSLPSLPSNSSQERPLDTRDPLLARAELALLSIVFVAVALSNGLVLAALARRGRRGHWAPIHVFIGHLCLADLAVALFQVLPQLAWKATDRFRGPDALCRAVKYLQMVGMYASSYMILAMTLDRHRAICRPMLAYRHGSGAHWNRPVLVAWAFSLLLSLPQLFIFAQRNVEGGSGVTDCWACFAEPWGRRTYVTWIALMVFVAPTLGIAACQVLIFREIHASLVPGPSERPGGRRRGRRTGSPGEGAHVSAAVAKTVRMTLVIVVVYVLCWAPFFLVQLWAAWDPEAPLEGAPFVLLMLLASLNSCTNPWIYASFSSSVSSELRSLLCCARGRTPPSLGPQDESCTTASSSLAKDTSS
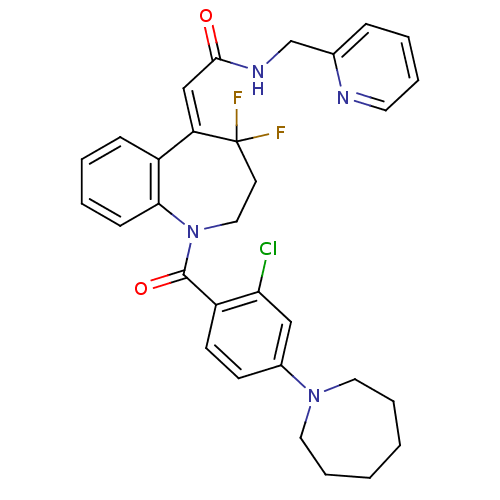
Inhibitor
Name:
BDBM35706
Synonyms:
5H-1-benzazepin-5-ylidene acetamide, 10m
Type:
Small organic molecule
Emp. Form.:
C31H31ClF2N4O2
Mol. Mass.:
565.053
SMILES:
FC1(F)CCN(C(=O)c2ccc(cc2Cl)N2CCCCCC2)c2ccccc2\C1=C\C(=O)NCc1ccccn1
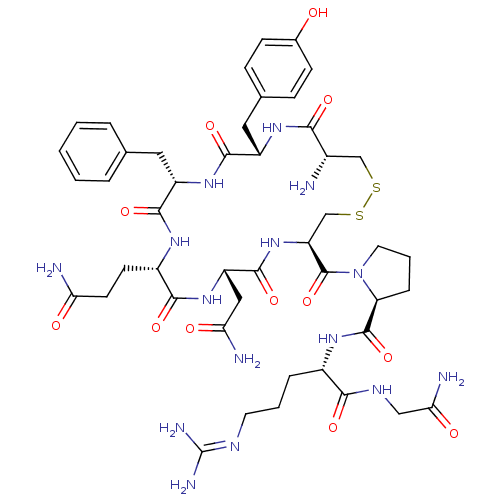
Substrate
Name:
BDBM35667
Synonyms:
AVP | CHEMBL373742 | US10131692, 44 (AVP) | [3H]Arginine vasopressin | [3H]Argipressin tannate | [3H]vasopressin
Type:
radiolabeled ligand
Emp. Form.:
C46H65N15O12S2
Mol. Mass.:
1084.232
SMILES:
[#7]-[#6@H]-1-[#6]-[#16]-[#16]-[#6]-[#6@H](-[#7]-[#6](=O)-[#6@H](-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6@H](-[#6]-c2ccccc2)-[#7]-[#6](=O)-[#6@H](-[#6]-c2ccc(-[#8])cc2)-[#7]-[#6]-1=O)-[#6](=O)-[#7]-1-[#6]-[#6]-[#6]-[#6@H]-1-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6](=O)-[#7]-[#6]-[#6](-[#7])=O