Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Glutamate receptor ionotropic, NMDA 1
Ligand
BDBM83449
Substrate
n/a
Meas. Tech.
ChEBML_154063
Ki
53±n/a nM
Citation
 Iorio, MA; Tomassini, L; Mattson, MV; George, C; Jacobson, AE Synthesis, stereochemistry, and biological activity of the 1-(1-phenyl-2-methylcyclohexyl)piperidines and the 1-(1-phenyl-4-methylcyclohexyl)piperidines. Absolute configuration of the potent trans-(-)-1-(1-phenyl-2-methylcyclohexyl)piperidine. J Med Chem 34:2615-23 (1991) [PubMed] Article
Iorio, MA; Tomassini, L; Mattson, MV; George, C; Jacobson, AE Synthesis, stereochemistry, and biological activity of the 1-(1-phenyl-2-methylcyclohexyl)piperidines and the 1-(1-phenyl-4-methylcyclohexyl)piperidines. Absolute configuration of the potent trans-(-)-1-(1-phenyl-2-methylcyclohexyl)piperidine. J Med Chem 34:2615-23 (1991) [PubMed] Article More Info.:
Target
Name:
Glutamate receptor ionotropic, NMDA 1
Synonyms:
Glutamate (NMDA) receptor subunit zeta 1 | Glutamate [NMDA] receptor subunit zeta-1 | Glutamate-NMDA-Channel | Glutamate-NMDA-MK801 | Glutamate-NMDA-Polyamine | Grin1 | NMDA | NMDZ1_RAT | Nmdar1 | phencyclidine
Type:
Enzyme Catalytic Domain
Mol. Mass.:
105533.40
Organism:
RAT
Description:
P35439
Residue:
938
Sequence:
MSTMHLLTFALLFSCSFARAACDPKIVNIGAVLSTRKHEQMFREAVNQANKRHGSWKIQLNATSVTHKPNAIQMALSVCEDLISSQVYAILVSHPPTPNDHFTPTPVSYTAGFYRIPVLGLTTRMSIYSDKSIHLSFLRTVPPYSHQSSVWFEMMRVYNWNHIILLVSDDHEGRAAQKRLETLLEERESKAEKVLQFDPGTKNVTALLMEARELEARVIILSASEDDAATVYRAAAMLNMTGSGYVWLVGEREISGNALRYAPDGIIGLQLINGKNESAHISDAVGVVAQAVHELLEKENITDPPRGCVGNTNIWKTGPLFKRVLMSSKYADGVTGRVEFNEDGDRKFANYSIMNLQNRKLVQVGIYNGTHVIPNDRKIIWPGGETEKPRGYQMSTRLKIVTIHQEPFVYVKPTMSDGTCKEEFTVNGDPVKKVICTGPNDTSPGSPRHTVPQCCYGFCIDLLIKLARTMNFTYEVHLVADGKFGTQERVNNSNKKEWNGMMGELLSGQADMIVAPLTINNERAQYIEFSKPFKYQGLTILVKKEIPRSTLDSFMQPFQSTLWLLVGLSVHVVAVMLYLLDRFSPFGRFKVNSEEEEEDALTLSSAMWFSWGVLLNSGIGEGAPRSFSARILGMVWAGFAMIIVASYTANLAAFLVLDRPEERITGINDPRLRNPSDKFIYATVKQSSVDIYFRRQVELSTMYRHMEKHNYESAAEAIQAVRDNKLHAFIWDSAVLEFEASQKCDLVTTGELFFRSGFGIGMRKDSPWKQNVSLSILKSHENGFMEDLDKTWVRYQECDSRSNAPATLTFENMAGVFMLVAGGIVAGIFLIFIEIAYKRHKDARRKQMQLAFAAVNVWRKNLQDRKSGRAEPDPKKKATFRAITSTLASSFKRRRSSKDTSTGGGRGALQNQKDTVLPRRAIEREEGQLQLCSRHRES
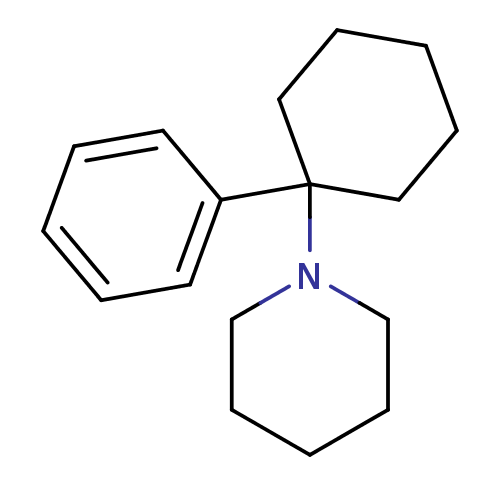
Inhibitor
Name:
BDBM83449
Synonyms:
1-(1-phenylcyclohexyl)piperidine;hydrochloride | MLS002320664 | PCP | PCP hydrochloride | PHENCYCLIDINE | Phencyclidine hydrochloride | SMR001338811 | cid_9795678
Type:
Small organic molecule
Emp. Form.:
C17H25N
Mol. Mass.:
243.3871
SMILES:
C1CCN(CC1)C1(CCCCC1)c1ccccc1