Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Prothrombin
Ligand
BDBM12658
Substrate
n/a
Meas. Tech.
ChEMBL_48307 (CHEMBL658196)
IC50
18±n/a nM
Citation
 Marlowe, CK; Sinha, U; Gunn, AC; Scarborough, RM Design, synthesis and structure-activity relationship of a series of arginine aldehyde factor Xa inhibitors. Part 1: structures based on the (D)-Arg-Gly-Arg tripeptide sequence. Bioorg Med Chem Lett 10:13-6 (2000) [PubMed] Article
Marlowe, CK; Sinha, U; Gunn, AC; Scarborough, RM Design, synthesis and structure-activity relationship of a series of arginine aldehyde factor Xa inhibitors. Part 1: structures based on the (D)-Arg-Gly-Arg tripeptide sequence. Bioorg Med Chem Lett 10:13-6 (2000) [PubMed] Article More Info.:
Target
Name:
Prothrombin
Synonyms:
Activation peptide fragment 1 | Activation peptide fragment 2 | Coagulation factor II | F2 | Prothrombin precursor | THRB_HUMAN | Thrombin heavy chain | Thrombin light chain
Type:
Protein
Mol. Mass.:
70029.57
Organism:
Homo sapiens (Human)
Description:
P00734
Residue:
622
Sequence:
MAHVRGLQLPGCLALAALCSLVHSQHVFLAPQQARSLLQRVRRANTFLEEVRKGNLERECVEETCSYEEAFEALESSTATDVFWAKYTACETARTPRDKLAACLEGNCAEGLGTNYRGHVNITRSGIECQLWRSRYPHKPEINSTTHPGADLQENFCRNPDSSTTGPWCYTTDPTVRRQECSIPVCGQDQVTVAMTPRSEGSSVNLSPPLEQCVPDRGQQYQGRLAVTTHGLPCLAWASAQAKALSKHQDFNSAVQLVENFCRNPDGDEEGVWCYVAGKPGDFGYCDLNYCEEAVEEETGDGLDEDSDRAIEGRTATSEYQTFFNPRTFGSGEADCGLRPLFEKKSLEDKTERELLESYIDGRIVEGSDAEIGMSPWQVMLFRKSPQELLCGASLISDRWVLTAAHCLLYPPWDKNFTENDLLVRIGKHSRTRYERNIEKISMLEKIYIHPRYNWRENLDRDIALMKLKKPVAFSDYIHPVCLPDRETAASLLQAGYKGRVTGWGNLKETWTANVGKGQPSVLQVVNLPIVERPVCKDSTRIRITDNMFCAGYKPDEGKRGDACEGDSGGPFVMKSPFNNRWYQMGIVSWGEGCDRDGKYGFYTHVFRLKKWIQKVIDQFGE
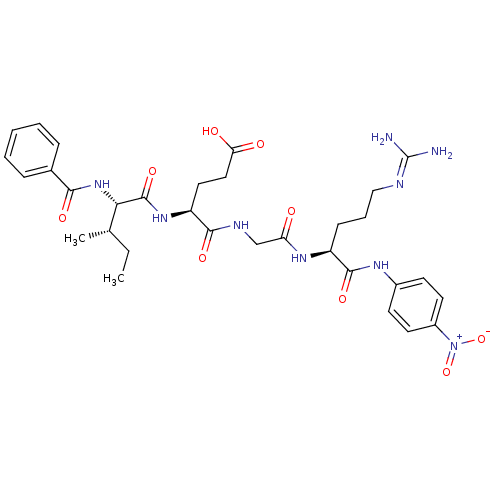
Inhibitor
Name:
BDBM12658
Synonyms:
4-[({[(1S)-4-carbamimidamido-1-[(4-nitrophenyl)carbamoyl]butyl]carbamoyl}methyl)carbamoyl]-4-[(2S,3S)-3-methyl-2-(phenylformamido)pentanamido]butanoic acid hydrochloride | Bz-Ile-Glu-Gly-Arg-pNA | Chromogenic Substrate S-2222 | L-Argininamide, N-benzoyl-L-isoleucyl-L-alpha-glutamylglycyl-N-(4-nitrophenyl)-, monohydrochloride | benzoyl-Ile-Glu-Gly-Arg-p-nitroanilide
Type:
Small organic molecule
Emp. Form.:
C32H43N9O9
Mol. Mass.:
697.7387
SMILES:
[#6]-[#6]-[#6@H](-[#6])-[#6@H](-[#7]-[#6](=O)-c1ccccc1)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#8])=O)-[#6](=O)-[#7]-[#6]-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6](=O)-[#7]-c1ccc(cc1)-[#7+](-[#8-])=O |r|