Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Glutamate receptor ionotropic, NMDA 1
Ligand
BDBM50123792
Substrate
n/a
Meas. Tech.
ChEMBL_140399 (CHEMBL746725)
IC50
1.5±n/a nM
Citation
 Katayama, S; Ae, N; Kodo, T; Masumoto, S; Hourai, S; Tamamura, C; Tanaka, H; Nagata, R Tricyclic indole-2-carboxylic acids: highly in vivo active and selective antagonists for the glycine binding site of the NMDA receptor. J Med Chem 46:691-701 (2003) [PubMed] Article
Katayama, S; Ae, N; Kodo, T; Masumoto, S; Hourai, S; Tamamura, C; Tanaka, H; Nagata, R Tricyclic indole-2-carboxylic acids: highly in vivo active and selective antagonists for the glycine binding site of the NMDA receptor. J Med Chem 46:691-701 (2003) [PubMed] Article More Info.:
Target
Name:
Glutamate receptor ionotropic, NMDA 1
Synonyms:
Glutamate (NMDA) receptor subunit zeta 1 | Glutamate [NMDA] receptor subunit zeta-1 | Glutamate-NMDA-Channel | Glutamate-NMDA-MK801 | Glutamate-NMDA-Polyamine | Grin1 | NMDA | NMDZ1_RAT | Nmdar1 | phencyclidine
Type:
Enzyme Catalytic Domain
Mol. Mass.:
105533.40
Organism:
RAT
Description:
P35439
Residue:
938
Sequence:
MSTMHLLTFALLFSCSFARAACDPKIVNIGAVLSTRKHEQMFREAVNQANKRHGSWKIQLNATSVTHKPNAIQMALSVCEDLISSQVYAILVSHPPTPNDHFTPTPVSYTAGFYRIPVLGLTTRMSIYSDKSIHLSFLRTVPPYSHQSSVWFEMMRVYNWNHIILLVSDDHEGRAAQKRLETLLEERESKAEKVLQFDPGTKNVTALLMEARELEARVIILSASEDDAATVYRAAAMLNMTGSGYVWLVGEREISGNALRYAPDGIIGLQLINGKNESAHISDAVGVVAQAVHELLEKENITDPPRGCVGNTNIWKTGPLFKRVLMSSKYADGVTGRVEFNEDGDRKFANYSIMNLQNRKLVQVGIYNGTHVIPNDRKIIWPGGETEKPRGYQMSTRLKIVTIHQEPFVYVKPTMSDGTCKEEFTVNGDPVKKVICTGPNDTSPGSPRHTVPQCCYGFCIDLLIKLARTMNFTYEVHLVADGKFGTQERVNNSNKKEWNGMMGELLSGQADMIVAPLTINNERAQYIEFSKPFKYQGLTILVKKEIPRSTLDSFMQPFQSTLWLLVGLSVHVVAVMLYLLDRFSPFGRFKVNSEEEEEDALTLSSAMWFSWGVLLNSGIGEGAPRSFSARILGMVWAGFAMIIVASYTANLAAFLVLDRPEERITGINDPRLRNPSDKFIYATVKQSSVDIYFRRQVELSTMYRHMEKHNYESAAEAIQAVRDNKLHAFIWDSAVLEFEASQKCDLVTTGELFFRSGFGIGMRKDSPWKQNVSLSILKSHENGFMEDLDKTWVRYQECDSRSNAPATLTFENMAGVFMLVAGGIVAGIFLIFIEIAYKRHKDARRKQMQLAFAAVNVWRKNLQDRKSGRAEPDPKKKATFRAITSTLASSFKRRRSSKDTSTGGGRGALQNQKDTVLPRRAIEREEGQLQLCSRHRES
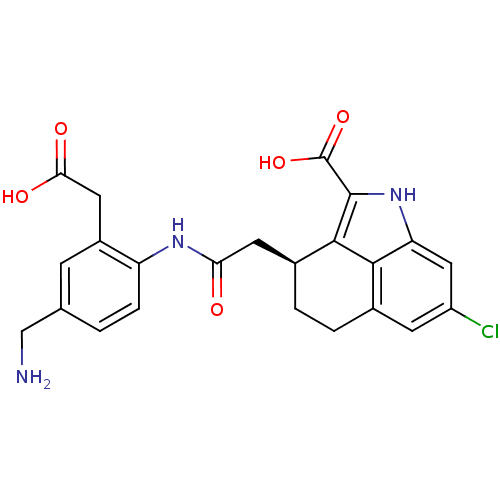
Inhibitor
Name:
BDBM50123792
Synonyms:
3-[(4-Aminomethyl-2-carboxymethyl-phenylcarbamoyl)-methyl]-7-chloro-1,3,4,5-tetrahydro-benzo[cd]indole-2-carboxylic acid | CHEMBL350974
Type:
Small organic molecule
Emp. Form.:
C23H22ClN3O5
Mol. Mass.:
455.891
SMILES:
NCc1ccc(NC(=O)C[C@@H]2CCc3cc(Cl)cc4[nH]c(C(O)=O)c2c34)c(CC(O)=O)c1