Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Aminopeptidase N
Ligand
BDBM50247539
Substrate
n/a
Meas. Tech.
ChEMBL_518900 (CHEMBL940644)
IC50
141900±n/a nM
Citation
More Info.:
Target
Name:
Aminopeptidase N
Synonyms:
AMPN_HUMAN | ANPEP | APN | Alanyl aminopeptidase | Aminopeptidase | CD13 | CD_antigen=CD13 | Microsomal aminopeptidase | Myeloid plasma membrane glycoprotein CD13 | PEPN | gp150 | hAPN
Type:
PROTEIN
Mol. Mass.:
109522.63
Organism:
Homo sapiens (Human)
Description:
ChEMBL_1507526
Residue:
967
Sequence:
MAKGFYISKSLGILGILLGVAAVCTIIALSVVYSQEKNKNANSSPVASTTPSASATTNPASATTLDQSKAWNRYRLPNTLKPDSYRVTLRPYLTPNDRGLYVFKGSSTVRFTCKEATDVIIIHSKKLNYTLSQGHRVVLRGVGGSQPPDIDKTELVEPTEYLVVHLKGSLVKDSQYEMDSEFEGELADDLAGFYRSEYMEGNVRKVVATTQMQAADARKSFPCFDEPAMKAEFNITLIHPKDLTALSNMLPKGPSTPLPEDPNWNVTEFHTTPKMSTYLLAFIVSEFDYVEKQASNGVLIRIWARPSAIAAGHGDYALNVTGPILNFFAGHYDTPYPLPKSDQIGLPDFNAGAMENWGLVTYRENSLLFDPLSSSSSNKERVVTVIAHELAHQWFGNLVTIEWWNDLWLNEGFASYVEYLGADYAEPTWNLKDLMVLNDVYRVMAVDALASSHPLSTPASEINTPAQISELFDAISYSKGASVLRMLSSFLSEDVFKQGLASYLHTFAYQNTIYLNLWDHLQEAVNNRSIQLPTTVRDIMNRWTLQMGFPVITVDTSTGTLSQEHFLLDPDSNVTRPSEFNYVWIVPITSIRDGRQQQDYWLIDVRAQNDLFSTSGNEWVLLNLNVTGYYRVNYDEENWRKIQTQLQRDHSAIPVINRAQIINDAFNLASAHKVPVTLALNNTLFLIEERQYMPWEAALSSLSYFKLMFDRSEVYGPMKNYLKKQVTPLFIHFRNNTNNWREIPENLMDQYSEVNAISTACSNGVPECEEMVSGLFKQWMENPNNNPIHPNLRSTVYCNAIAQGGEEEWDFAWEQFRNATLVNEADKLRAALACSKELWILNRYLSYTLNPDLIRKQDATSTIISITNNVIGQGLVWDFVQSNWKKLFNDYGGGSFSFSNLIQAVTRRFSTEYELQQLEQFKKDNEETGFGSGTRALEQALEKTKANIKWVKENKEVVLQWFTENSK
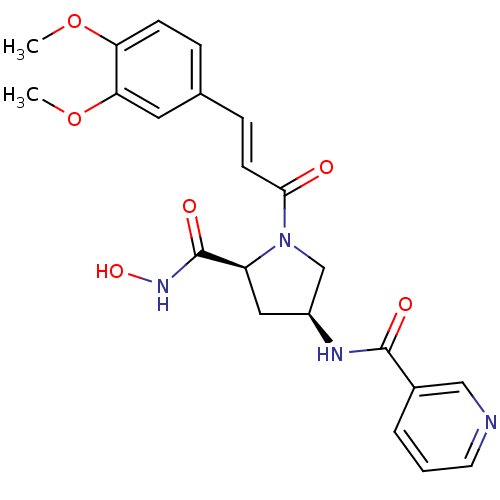
Inhibitor
Name:
BDBM50247539
Synonyms:
CHEMBL472915 | N-((3S,5S)-1-((E)-3-(3,4-dimethoxyphenyl)acryloyl)-5-(hydroxycarbamoyl)pyrrolidin-3-yl)nicotinamide | N-((3S,5S)-1-(3-(3,4-dimethoxyphenyl)acryloyl)-5-(hydroxycarbamoyl)pyrrolidin-3-yl)nicotinamide
Type:
Small organic molecule
Emp. Form.:
C22H24N4O6
Mol. Mass.:
440.4492
SMILES:
COc1ccc(\C=C\C(=O)N2C[C@H](C[C@H]2C(=O)NO)NC(=O)c2cccnc2)cc1OC |r|