Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Mitogen-activated protein kinase 10
Ligand
BDBM16016
Substrate
n/a
Meas. Tech.
ChEMBL_700502 (CHEMBL1648161)
IC50
1.6±n/a nM
Citation
 Probst, GD; Bowers, S; Sealy, JM; Truong, AP; Hom, RK; Galemmo, RA; Konradi, AW; Sham, HL; Quincy, DA; Pan, H; Yao, N; Lin, M; Tóth, G; Artis, DR; Zmolek, W; Wong, K; Qin, A; Lorentzen, C; Nakamura, DF; Quinn, KP; Sauer, JM; Powell, K; Ruslim, L; Wright, S; Chereau, D; Ren, Z; Anderson, JP; Bard, F; Yednock, TA; Griswold-Prenner, I Highly selective c-Jun N-terminal kinase (JNK) 2 and 3 inhibitors with in vitro CNS-like pharmacokinetic properties prevent neurodegeneration. Bioorg Med Chem Lett 21:315-9 (2010) [PubMed] Article
Probst, GD; Bowers, S; Sealy, JM; Truong, AP; Hom, RK; Galemmo, RA; Konradi, AW; Sham, HL; Quincy, DA; Pan, H; Yao, N; Lin, M; Tóth, G; Artis, DR; Zmolek, W; Wong, K; Qin, A; Lorentzen, C; Nakamura, DF; Quinn, KP; Sauer, JM; Powell, K; Ruslim, L; Wright, S; Chereau, D; Ren, Z; Anderson, JP; Bard, F; Yednock, TA; Griswold-Prenner, I Highly selective c-Jun N-terminal kinase (JNK) 2 and 3 inhibitors with in vitro CNS-like pharmacokinetic properties prevent neurodegeneration. Bioorg Med Chem Lett 21:315-9 (2010) [PubMed] Article More Info.:
Target
Name:
Mitogen-activated protein kinase 10
Synonyms:
JNK3 | JNK3A | MAP kinase p49 3F12 | MAPK10 | MK10_HUMAN | Mitogen-Activated Protein Kinase 10 (JNK3) | Mitogen-activated protein kinase 10 (Stress-activated protein kinase JNK3) (c-Jun N-terminal kinase 3) (MAP kinase p49 3F12) | Mitogen-activated protein kinase 10/Receptor-interacting serine/threonine-protein kinase 1 | PRKM10 | SAPK1B | Stress-activated protein kinase JNK3 | c-Jun N-terminal kinase 3 (JNK3)
Type:
Enzyme
Mol. Mass.:
52586.89
Organism:
Homo sapiens (Human)
Description:
n/a
Residue:
464
Sequence:
MSLHFLYYCSEPTLDVKIAFCQGFDKQVDVSYIAKHYNMSKSKVDNQFYSVEVGDSTFTVLKRYQNLKPIGSGAQGIVCAAYDAVLDRNVAIKKLSRPFQNQTHAKRAYRELVLMKCVNHKNIISLLNVFTPQKTLEEFQDVYLVMELMDANLCQVIQMELDHERMSYLLYQMLCGIKHLHSAGIIHRDLKPSNIVVKSDCTLKILDFGLARTAGTSFMMTPYVVTRYYRAPEVILGMGYKENVDIWSVGCIMGEMVRHKILFPGRDYIDQWNKVIEQLGTPCPEFMKKLQPTVRNYVENRPKYAGLTFPKLFPDSLFPADSEHNKLKASQARDLLSKMLVIDPAKRISVDDALQHPYINVWYDPAEVEAPPPQIYDKQLDEREHTIEEWKELIYKEVMNSEEKTKNGVVKGQPSPSGAAVNSSESLPPSSSVNDISSMSTDQTLASDTDSSLEASAGPLGCCR
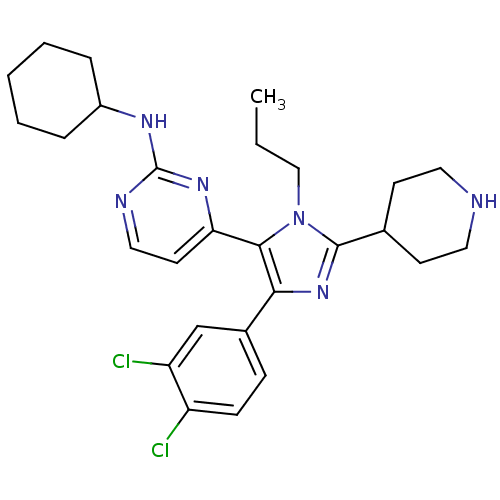
Inhibitor
Name:
BDBM16016
Synonyms:
CHEMBL437747 | N-cyclohexyl-4-[4-(3,4-dichlorophenyl)-2-(piperidin-4-yl)-1-propyl-1H-imidazol-5-yl]pyrimidin-2-amine | N-cyclohexyl-4-[4-(3,4-dichlorophenyl)-2-piperidin-4-yl-1-propyl-1H-imidazol-5-yl]pyrimidin-2-amine | imidazole-pyrimidine compound 2
Type:
Small organic molecule
Emp. Form.:
C27H34Cl2N6
Mol. Mass.:
513.505
SMILES:
CCCn1c(nc(c1-c1ccnc(NC2CCCCC2)n1)-c1ccc(Cl)c(Cl)c1)C1CCNCC1