Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Cholecystokinin receptor type A/Gastrin/cholecystokinin type B receptor
Ligand
BDBM50005463
Substrate
n/a
Meas. Tech.
ChEMBL_49424 (CHEMBL659627)
IC50
270±n/a nM
Citation
 Evans, BE; Rittle, KE; Bock, MG; DiPardo, RM; Freidinger, RM; Whitter, WL; Lundell, GF; Veber, DF; Anderson, PS; Chang, RS Methods for drug discovery: development of potent, selective, orally effective cholecystokinin antagonists. J Med Chem 31:2235-46 (1989) [PubMed] Article
Evans, BE; Rittle, KE; Bock, MG; DiPardo, RM; Freidinger, RM; Whitter, WL; Lundell, GF; Veber, DF; Anderson, PS; Chang, RS Methods for drug discovery: development of potent, selective, orally effective cholecystokinin antagonists. J Med Chem 31:2235-46 (1989) [PubMed] Article More Info.:
Target
Name:
Cholecystokinin receptor type A/Gastrin/cholecystokinin type B receptor
Synonyms:
Cholecystokinin receptor
Type:
n/a
Mol. Mass.:
n/a
Description:
ASSAY_ID of ChEMBL is 357790
Components:
This complex has 2 components.
Component 1
Name:
Cholecystokinin receptor type A
Synonyms:
CCK-A receptor | CCK-AR | CCK1-R | CCKAR | CCKAR_HUMAN | CCKRA | Cholecystokinin receptor | Cholecystokinin receptor type A | Cholecystokinin-1 Receptor
Type:
G Protein-Coupled Receptor (GPCR)
Mol. Mass.:
47859.34
Organism:
Homo sapiens (Human)
Description:
Stable expression of human CCK-1 receptors in HEK 293 cells.
Residue:
428
Sequence:
MDVVDSLLVNGSNITPPCELGLENETLFCLDQPRPSKEWQPAVQILLYSLIFLLSVLGNTLVITVLIRNKRMRTVTNIFLLSLAVSDLMLCLFCMPFNLIPNLLKDFIFGSAVCKTTTYFMGTSVSVSTFNLVAISLERYGAICKPLQSRVWQTKSHALKVIAATWCLSFTIMTPYPIYSNLVPFTKNNNQTANMCRFLLPNDVMQQSWHTFLLLILFLIPGIVMMVAYGLISLELYQGIKFEASQKKSAKERKPSTTSSGKYEDSDGCYLQKTRPPRKLELRQLSTGSSSRANRIRSNSSAANLMAKKRVIRMLIVIVVLFFLCWMPIFSANAWRAYDTASAERRLSGTPISFILLLSYTSSCVNPIIYCFMNKRFRLGFMATFPCCPNPGPPGARGEVGEEEEGGTTGASLSRFSYSHMSASVPPQ
Component 2
Name:
Gastrin/cholecystokinin type B receptor
Synonyms:
CCK-2 receptor | CCK-B receptor | CCK-BR | CCKBR | CCKRB | Cholecystokinin A | Cholecystokinin receptor | Cholecystokinin-2 Receptor | GASR_HUMAN | Gastrin/cholecystokinin type B receptor
Type:
G Protein-Coupled Receptor (GPCR)
Mol. Mass.:
48445.79
Organism:
Homo sapiens (Human)
Description:
Stable expression of human CCK-2 receptors in HEK 293 cells.
Residue:
447
Sequence:
MELLKLNRSVQGTGPGPGASLCRPGAPLLNSSSVGNLSCEPPRIRGAGTRELELAIRITLYAVIFLMSVGGNMLIIVVLGLSRRLRTVTNAFLLSLAVSDLLLAVACMPFTLLPNLMGTFIFGTVICKAVSYLMGVSVSVSTLSLVAIALERYSAICRPLQARVWQTRSHAARVIVATWLLSGLLMVPYPVYTVVQPVGPRVLQCVHRWPSARVRQTWSVLLLLLLFFIPGVVMAVAYGLISRELYLGLRFDGDSDSDSQSRVRNQGGLPGAVHQNGRCRPETGAVGEDSDGCYVQLPRSRPALELTALTAPGPGSGSRPTQAKLLAKKRVVRMLLVIVVLFFLCWLPVYSANTWRAFDGPGAHRALSGAPISFIHLLSYASACVNPLVYCFMHRRFRQACLETCARCCPRPPRARPRALPDEDPPTPSIASLSRLSYTTISTLGPG
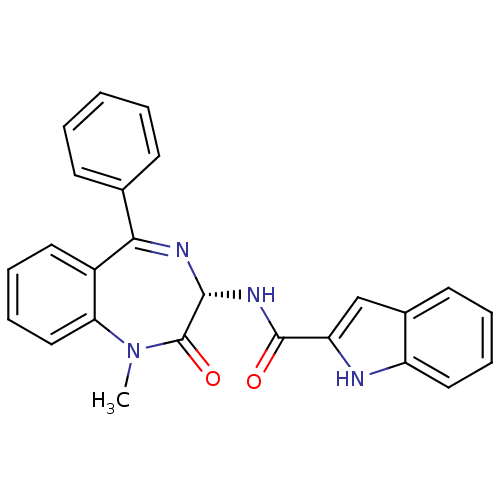
Inhibitor
Name:
BDBM50005463
Synonyms:
(R)-1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide: 0.1C4H10O | (S)-1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide | (S)-N-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-1H-indole-2-carboxamide | (Z)-N-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-1H-indole-2-carboxamide | 1H-Indole-2-carboxylic acid ((S)-1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide | 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide | 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide (L-364,718 ((S)-devazepide) | 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide (MK-329, L-364,718) | 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(Devazepide or (R) L364718) | 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(L-364718) | 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(devazepide) | 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide: 0.1C4H10O. 0.15CH2Cl2 | CCK antagonist synthetic 17 | CCK antagonist synthetic 18 | CHEMBL9506 | DEVAZEPIDE | L-364,718 | L-364718 | MK-329
Type:
Small organic molecule
Emp. Form.:
C25H20N4O2
Mol. Mass.:
408.4519
SMILES:
CN1c2ccccc2C(=N[C@H](NC(=O)c2cc3ccccc3[nH]2)C1=O)c1ccccc1 |r,c:9|