Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Neuromedin-K receptor
Ligand
BDBM50299467
Substrate
n/a
Meas. Tech.
ChEMBL_883638 (CHEMBL2210303)
EC50
0.71±n/a nM
Citation
 Bissantz, C; Bohnert, C; Hoffmann, T; Marcuz, A; Schnider, P; Malherbe, P Identification of a crucial amino acid in the helix position 6.51 of human tachykinin neurokinin 1 and 3 receptors contributing to the insurmountable mode of antagonism by dual NK1/NK3 antagonists. J Med Chem 55:5061-76 (2012) [PubMed] Article
Bissantz, C; Bohnert, C; Hoffmann, T; Marcuz, A; Schnider, P; Malherbe, P Identification of a crucial amino acid in the helix position 6.51 of human tachykinin neurokinin 1 and 3 receptors contributing to the insurmountable mode of antagonism by dual NK1/NK3 antagonists. J Med Chem 55:5061-76 (2012) [PubMed] Article More Info.:
Target
Name:
Neuromedin-K receptor
Synonyms:
NK-3 receptor | NK-3R | NK3R | NK3R_HUMAN | NKR | Neurokinin 3 receptor | Neurokinin B receptor | Neurokinin-3 (NK-3) | Neuromedin-3 receptor (NK-3R) | Neuromedin-3 receptor (NK3) | Neuromedin-K receptor | Neuromedin-K receptor (NK-3 receptor) | Neuromedin-K receptor (NK3) | Neuromedin-K receptor(NK3R) | TAC3R | TACR3 | Tachykinin receptor 3 | Tachykinin receptor 3 (NK3)
Type:
Enzyme
Mol. Mass.:
52221.96
Organism:
Homo sapiens (Human)
Description:
P29371
Residue:
465
Sequence:
MATLPAAETWIDGGGGVGADAVNLTASLAAGAATGAVETGWLQLLDQAGNLSSSPSALGLPVASPAPSQPWANLTNQFVQPSWRIALWSLAYGVVVAVAVLGNLIVIWIILAHKRMRTVTNYFLVNLAFSDASMAAFNTLVNFIYALHSEWYFGANYCRFQNFFPITAVFASIYSMTAIAVDRYMAIIDPLKPRLSATATKIVIGSIWILAFLLAFPQCLYSKTKVMPGRTLCFVQWPEGPKQHFTYHIIVIILVYCFPLLIMGITYTIVGITLWGGEIPGDTCDKYHEQLKAKRKVVKMMIIVVMTFAICWLPYHIYFILTAIYQQLNRWKYIQQVYLASFWLAMSSTMYNPIIYCCLNKRFRAGFKRAFRWCPFIKVSSYDELELKTTRFHPNRQSSMYTVTRMESMTVVFDPNDADTTRSSRKKRATPRDPSFNGCSRRNSKSASATSSFISSPYTSVDEYS
Inhibitor
Name:
BDBM50299467
Synonyms:
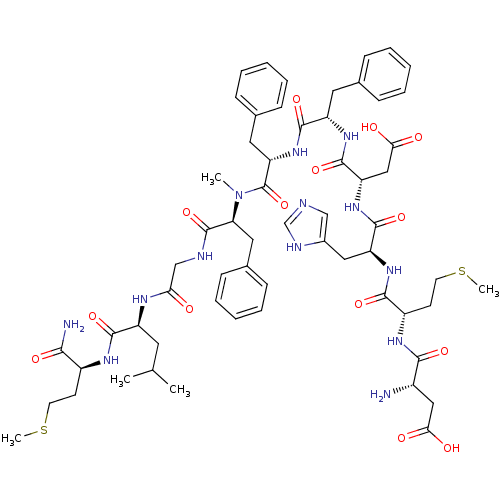
(5S,8S,14S,17S,20S,23S,26S,29S,32S)-26-((1H-imidazol-5-yl)methyl)-32-amino-14,17,20-tribenzyl-5-carbamoyl-23-(carboxymethyl)-8-isobutyl-15-methyl-29-(2-(methylthio)ethyl)-7,10,13,16,19,22,25,28,31-nonaoxo-2-thia-6,9,12,15,18,21,24,27,30-nonaazatetratriacontan-34-oic acid | CHEMBL583102
Type:
Small organic molecule
Emp. Form.:
C60H81N13O14S2
Mol. Mass.:
1272.494
SMILES:
CSCC[C@H](NC(=O)[C@H](CC(C)C)NC(=O)CNC(=O)[C@H](Cc1ccccc1)N(C)C(=O)[C@H](Cc1ccccc1)NC(=O)[C@H](Cc1ccccc1)NC(=O)[C@H](CC(O)=O)NC(=O)[C@H](Cc1cnc[nH]1)NC(=O)[C@H](CCSC)NC(=O)[C@@H](N)CC(O)=O)C(N)=O |r|