Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
D(3) dopamine receptor
Ligand
BDBM50007518
Substrate
n/a
Ki
0.1±n/a nM
Comments
PDSP_803
Citation
 Toll, L; Berzetei-Gurske, IP; Polgar, WE; Brandt, SR; Adapa, ID; Rodriguez, L; Schwartz, RW; Haggart, D; O'Brien, A; White, A; Kennedy, JM; Craymer, K; Farrington, L; Auh, JS Standard binding and functional assays related to medications development division testing for potential cocaine and opiate narcotic treatment medications. NIDA Res Monogr 178:440-66 (1998) [PubMed]
Toll, L; Berzetei-Gurske, IP; Polgar, WE; Brandt, SR; Adapa, ID; Rodriguez, L; Schwartz, RW; Haggart, D; O'Brien, A; White, A; Kennedy, JM; Craymer, K; Farrington, L; Auh, JS Standard binding and functional assays related to medications development division testing for potential cocaine and opiate narcotic treatment medications. NIDA Res Monogr 178:440-66 (1998) [PubMed] More Info.:
Target
Name:
D(3) dopamine receptor
Synonyms:
DOPAMINE D3 | DRD3 | DRD3_HUMAN | Dopamine D3 receptor | Dopamine D3 receptor (D3) | Dopamine D3 receptor (D3R) | Dopamine receptor | Dopamine receptor (D3) | Dopamine receptor D3
Type:
n/a
Mol. Mass.:
44243.43
Organism:
Homo sapiens (Human)
Description:
n/a
Residue:
400
Sequence:
MASLSQLSSHLNYTCGAENSTGASQARPHAYYALSYCALILAIVFGNGLVCMAVLKERALQTTTNYLVVSLAVADLLVATLVMPWVVYLEVTGGVWNFSRICCDVFVTLDVMMCTASILNLCAISIDRYTAVVMPVHYQHGTGQSSCRRVALMITAVWVLAFAVSCPLLFGFNTTGDPTVCSISNPDFVIYSSVVSFYLPFGVTVLVYARIYVVLKQRRRKRILTRQNSQCNSVRPGFPQQTLSPDPAHLELKRYYSICQDTALGGPGFQERGGELKREEKTRNSLSPTIAPKLSLEVRKLSNGRLSTSLKLGPLQPRGVPLREKKATQMVAIVLGAFIVCWLPFFLTHVLNTHCQTCHVSPELYSATTWLGYVNSALNPVIYTTFNIEFRKAFLKILSC
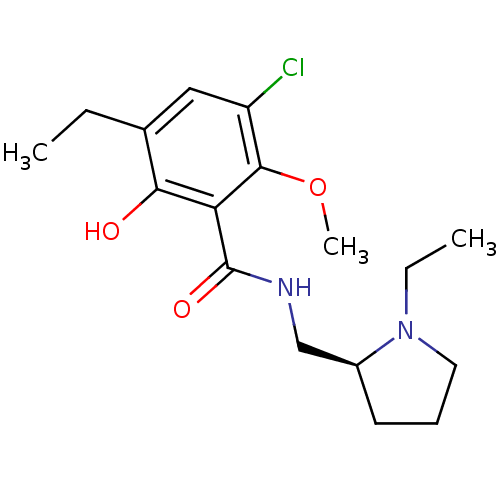
Inhibitor
Name:
BDBM50007518
Synonyms:
(S)-3-chloro-5-ethyl-N-((1-ethylpyrrolidin-2-yl)methyl)-6-hydroxy-2-methoxybenzamide | 3,5-Dichloro-N-(1-ethyl-pyrrolidin-2-ylmethyl)-2-hydroxy-6-methoxy-benzamide | 3-Chloro-5-ethyl-N-((S)-1-ethyl-pyrrolidin-2-ylmethyl)-6-hydroxy-2-methoxy-benzamide | 3-Chloro-5-ethyl-N-(1-ethyl-pyrrolidin-2-ylmethyl)-6-hydroxy-2-methoxy-benzamide | 3-Chloro-5-ethyl-N-(1-ethyl-pyrrolidin-2-ylmethyl)-6-hydroxy-2-methoxy-benzamide (etichlopride) | 3-Chloro-5-ethyl-N-(1-ethyl-pyrrolidin-2-ylmethyl)-6-hydroxy-2-methoxy-benzamide(Eticlopride) | 3-chloro-5-ethyl-N-{[(2S)-1-ethylpyrrolidin-2-yl]methyl}-6-hydroxy-2-methoxybenzamide | CHEMBL8946 | ETICLOPRIDE | ETICLOPRIDE,S(-) | Eticlopride;3-Chloro-5-ethyl-N-(1-ethyl-pyrrolidin-2-ylmethyl)-6-hydroxy-2-methoxy-benzamide
Type:
Small organic molecule
Emp. Form.:
C17H25ClN2O3
Mol. Mass.:
340.845
SMILES:
CCN1CCC[C@H]1CNC(=O)c1c(O)c(CC)cc(Cl)c1OC