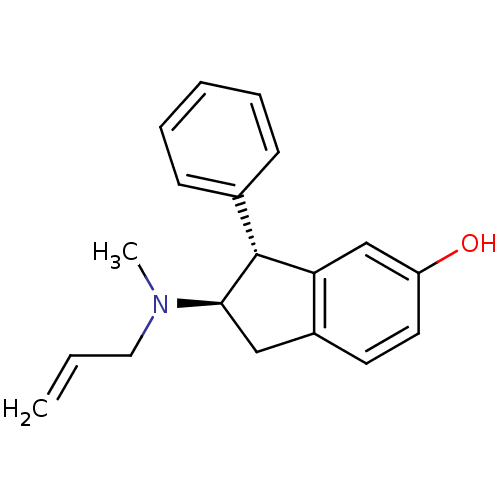
BDBM50054084 (2R,3R)-2-(Allyl-methyl-amino)-3-phenyl-indan-5-ol::CHEMBL135197
SMILES CN(CC=C)[C@@H]1Cc2ccc(O)cc2[C@H]1c1ccccc1
InChI Key InChIKey=XWQIFHFKVSCKAY-RTBURBONSA-N
Data 4 KI
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 50054084
Found 4 hits for monomerid = 50054084
Affinity DataKi: 0.430nMAssay Description:Binding affinity at Dopamine receptor D2 in rat striatal homogenate by [3H]-spiperone displacement.More data for this Ligand-Target Pair
Affinity DataKi: 0.430nMAssay Description:Binding affinity at Dopamine receptor D2 in rat striatal homogenate by [3H]-spiperone displacement.More data for this Ligand-Target Pair
Affinity DataKi: 0.790nMAssay Description:Binding affinity at Dopamine receptor D1 in rat neostriatum by [3H]-SCH- 23390 displacement.More data for this Ligand-Target Pair
Affinity DataKi: 0.790nMAssay Description:Binding affinity at Dopamine receptor D1 in rat neostriatum by [3H]-SCH- 23390 displacement.More data for this Ligand-Target Pair