Discovery of Novel Peptidomimetic Boronate ClpP Inhibitors with Noncanonical Enzyme Mechanism as Potent Virulence Blockersin Vitroandin Vivo.
Ju, Y., He, L., Zhou, Y., Yang, T., Sun, K., Song, R., Yang, Y., Li, C., Sang, Z., Bao, R., Luo, Y.(2020) J Med Chem 63: 3104-3119
- PubMed: 32031798
- DOI: https://doi.org/10.1021/acs.jmedchem.9b01746
- Primary Citation of Related Structures:
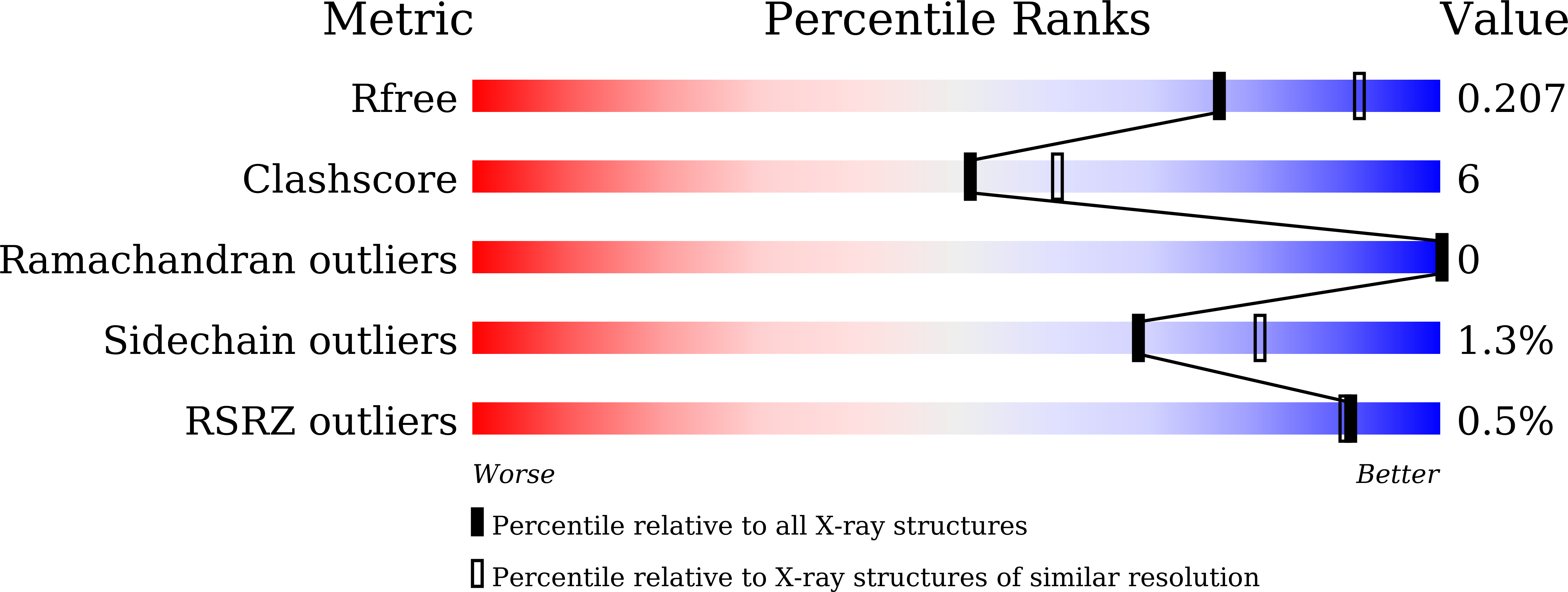
6L3X, 6L40 - PubMed Abstract:
Caseinolytic protease P (ClpP) is considered as a promising target for the treatment of Staphylococcus aureus infections. In an unbiased screen of 2632 molecules, a peptidomimetic boronate, MLN9708, was found to be a potent suppressor of Sa ClpP function. A time-saving and cost-efficient strategy integrating in silico position scanning, multistep miniaturized synthesis, and bioactivity testing was deployed for optimization of this hit compound and led to fast exploration of structure-activity relationships. Five of 150 compounds from the miniaturized synthesis exhibited improved inhibitory activity. Compound 43Hf was the most active inhibitor and showed reversible covalent binding to Sa ClpP while did not destabilize the tetradecameric structure of Sa ClpP. The crystal structure of 43Hf - Sa ClpP complex provided mechanistic insight into the covalent binding mode of peptidomimetic boronate and Sa ClpP. Furthermore, 43Hf could bind endogenous ClpP in S. aureus cells and exhibited significant efficacy in attenuating S. aureus virulence in vitro and in vivo .
Organizational Affiliation:
State Key Laboratory of Biotherapy and Cancer Center/Collaborative Innovation Center for Biotherapy, West China Hospital, Sichuan University, Chengdu 610041, China.