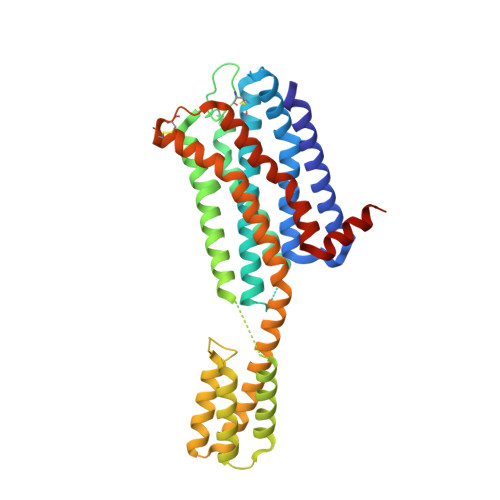
Structure of the dopamine D 2 receptor in complex with the antipsychotic drug spiperone.
Im, D., Inoue, A., Fujiwara, T., Nakane, T., Yamanaka, Y., Uemura, T., Mori, C., Shiimura, Y., Kimura, K.T., Asada, H., Nomura, N., Tanaka, T., Yamashita, A., Nango, E., Tono, K., Kadji, F.M.N., Aoki, J., Iwata, S., Shimamura, T.(2020) Nat Commun 11: 6442-6442
- PubMed: 33353947
- DOI: https://doi.org/10.1038/s41467-020-20221-0
- Primary Citation of Related Structures:
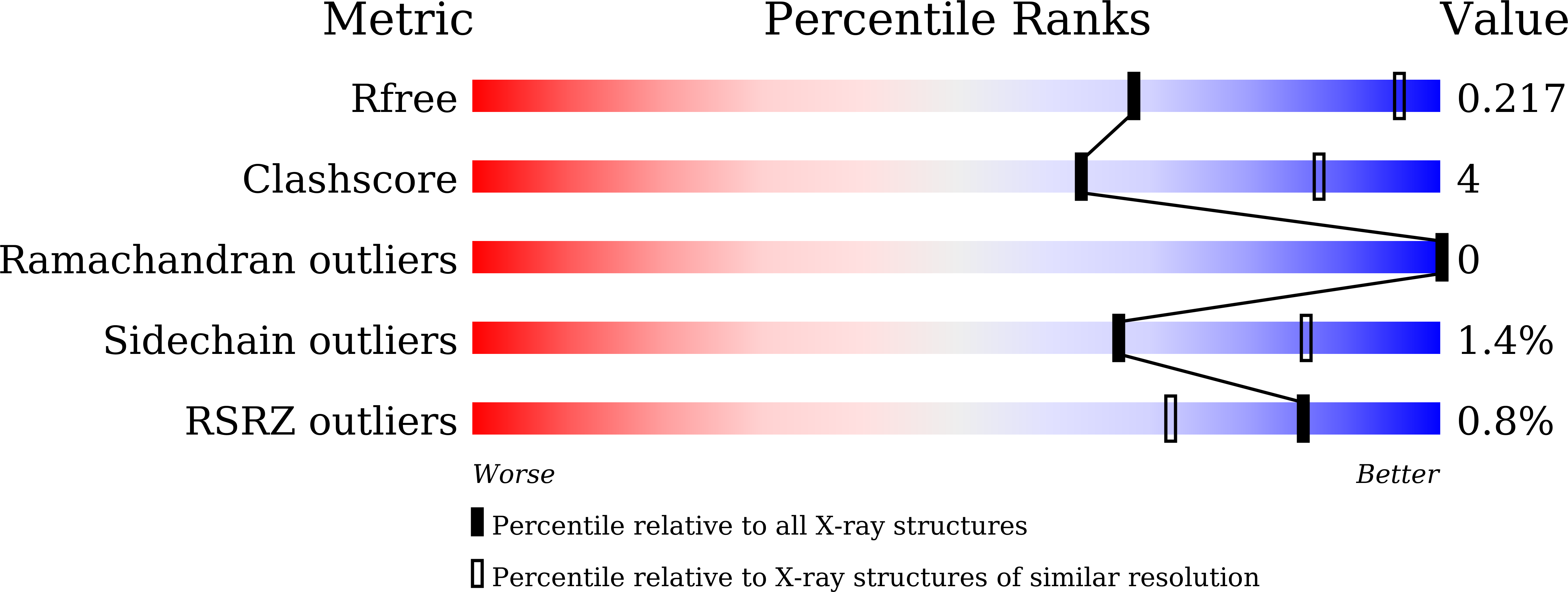
7DFP - PubMed Abstract:
In addition to the serotonin 5-HT 2A receptor (5-HT 2A R), the dopamine D 2 receptor (D 2 R) is a key therapeutic target of antipsychotics for the treatment of schizophrenia. The inactive state structures of D 2 R have been described in complex with the inverse agonists risperidone (D 2 R ris ) and haloperidol (D 2 R hal ). Here we describe the structure of human D 2 R in complex with spiperone (D 2 R spi ). In D 2 R spi , the conformation of the extracellular loop (ECL) 2, which composes the ligand-binding pocket, was substantially different from those in D 2 R ris and D 2 R hal , demonstrating that ECL2 in D 2 R is highly dynamic. Moreover, D 2 R spi exhibited an extended binding pocket to accommodate spiperone's phenyl ring, which probably contributes to the selectivity of spiperone to D 2 R and 5-HT 2A R. Together with D 2 R ris and D 2 R hal , the structural information of D 2 R spi should be of value for designing novel antipsychotics with improved safety and efficacy.
Organizational Affiliation:
Department of Cell Biology, Graduate School of Medicine, Kyoto University, Kyoto, Japan.