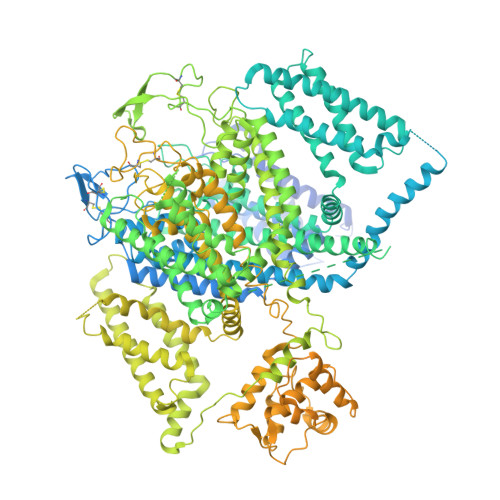
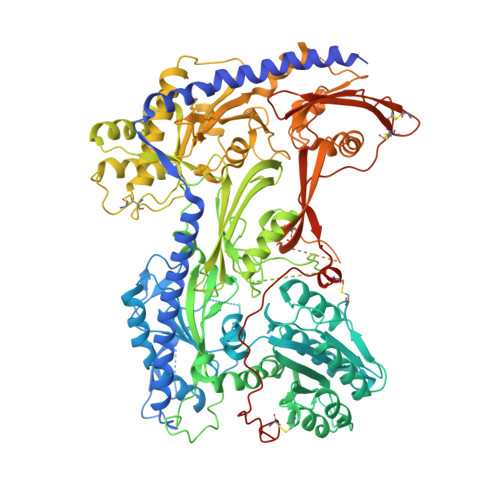
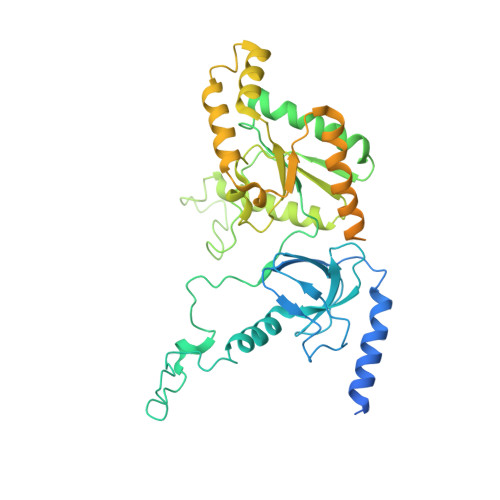
Structural basis for pore blockade of human voltage-gated calcium channel Ca v 1.3 by motion sickness drug cinnarizine.
Yao, X., Gao, S., Yan, N.(2022) Cell Res 32: 946-948
- PubMed: 35477996
- DOI: https://doi.org/10.1038/s41422-022-00663-5
- Primary Citation of Related Structures:
7UHF, 7UHG
Organizational Affiliation:
Department of Molecular Biology, Princeton University, Princeton, NJ, USA. xiayao@princeton.edu.