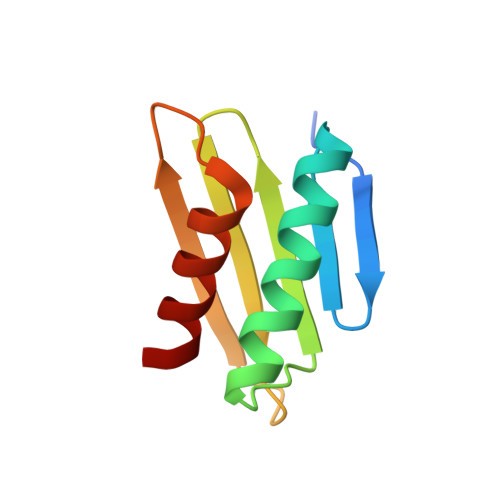
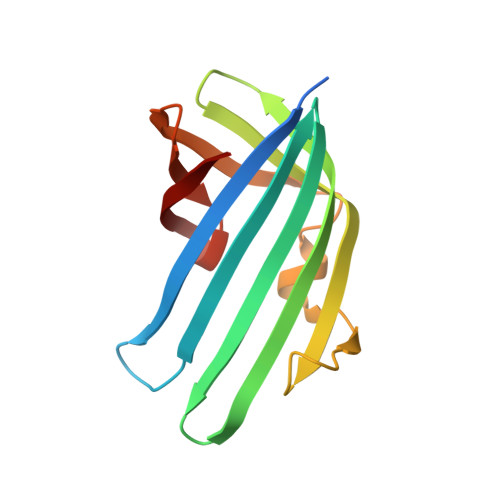
Bile salt receptor complex activates a pathogenic type III secretion system.
Li, P., Rivera-Cancel, G., Kinch, L.N., Salomon, D., Tomchick, D.R., Grishin, N.V., Orth, K.(2016) Elife 5
- PubMed: 27377244
- DOI: https://doi.org/10.7554/eLife.15718
- Primary Citation of Related Structures:
5KEV, 5KEW - PubMed Abstract:
Bile is an important component of the human gastrointestinal tract with an essential role in food absorption and antimicrobial activities. Enteric bacterial pathogens have developed strategies to sense bile as an environmental cue to regulate virulence genes during infection. We discovered that Vibrio parahaemolyticus VtrC, along with VtrA and VtrB, are required for activating the virulence type III secretion system 2 in response to bile salts. The VtrA/VtrC complex activates VtrB in the presence of bile salts. The crystal structure of the periplasmic domains of the VtrA/VtrC heterodimer reveals a β-barrel with a hydrophobic inner chamber. A co-crystal structure of VtrA/VtrC with bile salt, along with biophysical and mutational analysis, demonstrates that the hydrophobic chamber binds bile salts and activates the virulence network. As part of a family of conserved signaling receptors, VtrA/VtrC provides structural and functional insights into the evolutionarily conserved mechanism used by bacteria to sense their environment.
Organizational Affiliation:
Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, United States.