Studies on spiro[4.5]decanone prolyl hydroxylase domain inhibitors.
Holt-Martyn, J.P., Tumber, A., Rahman, M.Z., Lippl, K., Figg Jr., W., McDonough, M.A., Chowdhury, R., Schofield, C.J.(2019) Medchemcomm 10: 500-504
- PubMed: 31057728
- DOI: https://doi.org/10.1039/c8md00548f
- Primary Citation of Related Structures:
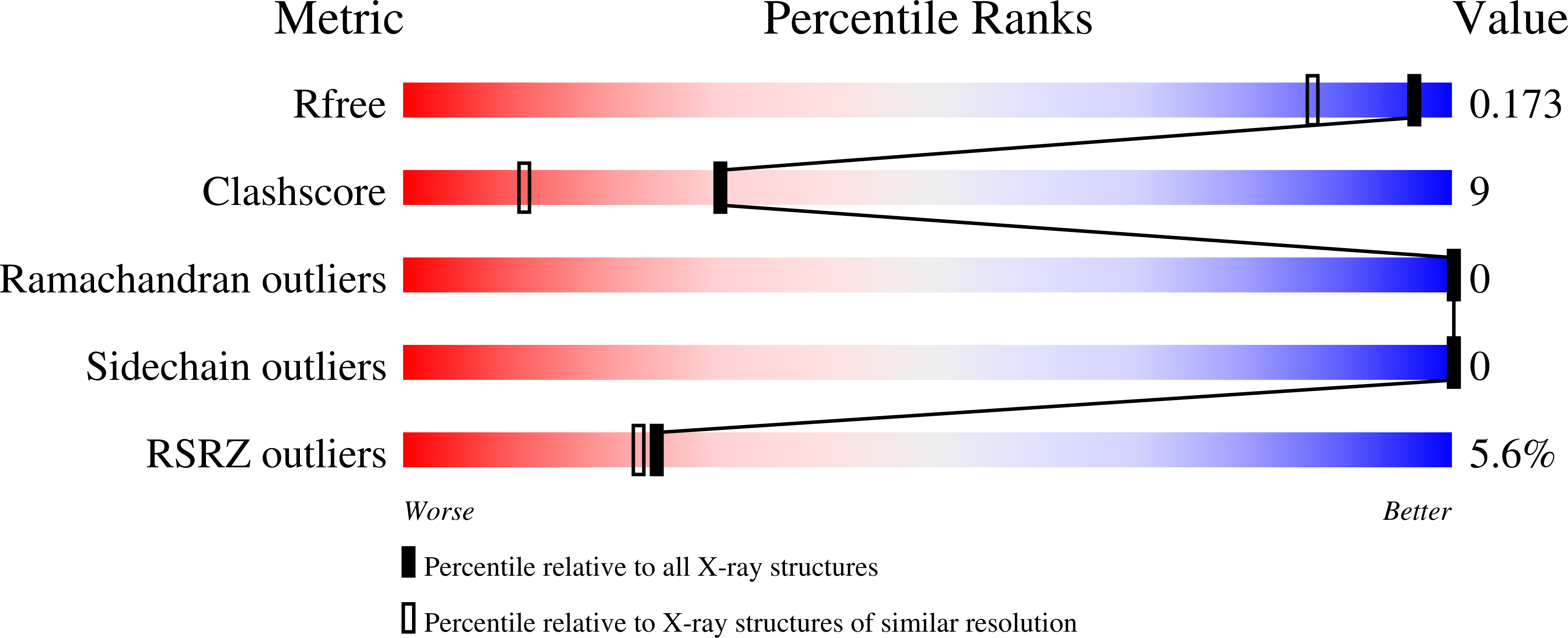
6QGV - PubMed Abstract:
The 2-oxoglutarate (2OG) dependent hypoxia inducible factor (HIF) prolyl hydroxylases (PHDs) are targets for treatment of anaemia and other ischaemia related diseases. PHD inhibitors are in clinical trials; however, the number of reported templates for PHD inhibition is limited. We report structure-activity relationship and crystallographic studies on spiro[4.5]decanone containing PHD inhibitors. Together with other studies, our results reveal spiro[4.5]decanones as useful templates for generation of potent and selective 2OG oxygenase inhibitors.
Organizational Affiliation:
Department of Chemistry , University of Oxford , Chemistry Research Laboratory , 12 Mansfield Road , Oxford , OX1 3TA , UK . Email: christopher.schofield@chem.ox.ac.uk.